Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals

Diseases articles from across Nature Portfolio
Diseases are abnormal conditions that have a specific set of signs and symptoms. Diseases can have an external cause, such as an infection, or an internal cause, such as autoimmune diseases.
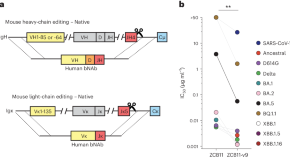
Affinity maturation of CRISPR-engineered B cell receptors in vivo
CRISPR–Cas12a was used to directly replace mouse antibody variable chain genes with human versions in primary B cells. The edited cells underwent affinity maturation in vivo, improving the potency of HIV-1 and SARS-CoV-2 neutralizing antibodies without loss of bioavailability. Affinity maturation of edited cells also enables new vaccine models and adaptive B cell therapies.
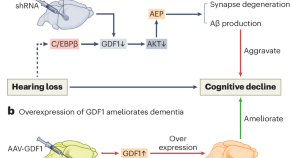
Hearing loss promotes Alzheimer’s disease
Epidemiological studies reveal a correlation between hearing loss and the development and progression of Alzheimer’s disease (AD), but the underlying causal mechanisms remain unclear. A study now provides experimental evidence that hearing loss can promote AD via the growth differentiation factor 1 (GDF1) pathway, which may aid in developing potential AD therapeutic strategies.
- Hong-Bo Zhao
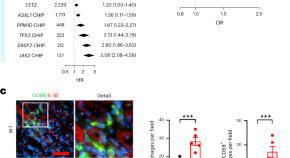
Acquired blood mutations cause acute kidney injury via dysregulated inflammation
Acute kidney injury affects one in five hospitalized patients and can lead to lasting kidney damage or death. We show that clonal hematopoiesis of indeterminate potential — a common age-related condition caused by blood cell mutations — increases the risk of acute kidney injury in multiple cohorts of human patients and in mouse models.
Related Subjects
- Cardiovascular diseases
- Dental diseases
- Endocrine system and metabolic diseases
- Eye diseases
- Gastrointestinal diseases
- Haematological diseases
- Immunological disorders
- Infectious diseases
- Kidney diseases
- Metabolic disorders
- Neurological disorders
- Nutrition disorders
- Oral diseases
- Psychiatric disorders
- Reproductive disorders
- Respiratory tract diseases
- Rheumatic diseases
- Skin diseases
- Urogenital diseases
Latest Research and Reviews
Invasive candidiasis
This PrimeView highlights the diagnosis of invasive candidiasis, and summarizes its epidemiology, mechanisms and management. It accompanies the Primer article on this topic by Lass- Flörl et al.
Is phase angle associated with visceral adiposity and cardiometabolic risk in cardiology outpatients?
- Victoria Domingues Ferraz
- Jarson Pedro da Costa Pereira
- Ilma Kruze Grande de Arruda
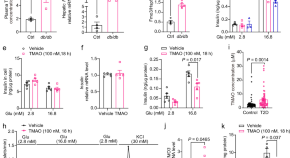
Trimethylamine N-oxide impairs β-cell function and glucose tolerance
β-Cell dysfunction is a hallmark of type 2 diabetes. Here, the authors show that trimethylamine N-oxide (TMAO (a microbiota metabolite)) induces β-cell dysfunction and type 2 diabetes in mice through NLRP3 inflammasome activation and calcium transients.
- Lijuan Kong
- Pingping Li
Invasive candidiasis is a fungal disease caused by Candida pathogens. In this Primer, Lass-Flörl et al. summarize current knowledge of epidemiology and mechanisms and discuss the diagnosis, treatment and quality of life of patients with invasive candidiasis. They also highlight outstanding research questions.
- Cornelia Lass-Flörl
- Souha S. Kanj
- Miriam Alisa Govrins
Incidence and prognosis of cardiac conduction system diseases in hypertension: the STEP trial
Zhao, Deng and colleagues present a post hoc analysis of the STEP trial showing that intensive blood pressure control does not reduce the risk of cardiac conduction diseases in older adults with hypertension.
Shipping-related pollution decreased but mortality increased in Chinese port cities
This study looks at the changes in Chinese port cities in relation to demography and emissions reductions to examine the relationship between health and emissions. They found that even though shipping-related PM 2.5 decreased, mortality associated with long-term exposure to it increased by 11%.
- Zhaofeng Lv
News and Comment
Comment on: current opinions on the management of prolonged ischemic priapism: does penoscrotal decompression outperform corporoglanular tunneling.
- Omer A. Raheem
- Marwan Alkassis
- Rosemary Iwuala
Cutting-edge CAR-T cancer therapy is now made in India — at one-tenth the cost
The treatment, called NexCAR19, raises hopes that this transformative class of medicine will become more readily available in low- and middle-income countries.
- Smriti Mallapaty
A new standard of care for advanced-stage urothelial carcinoma
- Peter Sidaway

AI hears hidden X factor in zebra finch love songs
Machine learning detects song differences too subtle for humans to hear, and physicists harness the computing power of the strange skyrmion.
- Nick Petrić Howe
- Benjamin Thompson
Correspondence to “Relationship between employment and quality of life and self-perceived health in people with spinal cord injury: an international comparative study based on the InSCI community survey”
Salivary lipid changes in young adult tobacco smokers and e-cigarette users: a hidden risk to oral health.
- Omer Waleed Majid
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
- Open access
- Published: 18 November 2021
Scientific evidence based rare disease research discovery with research funding data in knowledge graph
- Qian Zhu ORCID: orcid.org/0000-0002-4858-6333 1 ,
- Ðắc-Trung Nguyễn 1 ,
- Timothy Sheils 1 ,
- Gioconda Alyea 2 ,
- Eric Sid 3 ,
- Yanji Xu 3 ,
- James Dickens 3 ,
- Ewy A. Mathé 1 &
- Anne Pariser 3
Orphanet Journal of Rare Diseases volume 16 , Article number: 483 ( 2021 ) Cite this article
3173 Accesses
7 Citations
Metrics details
Limited knowledge and unclear underlying biology of many rare diseases pose significant challenges to patients, clinicians, and scientists. To address these challenges, there is an urgent need to inspire and encourage scientists to propose and pursue innovative research studies that aim to uncover the genetic and molecular causes of more rare diseases and ultimately to identify effective therapeutic solutions. A clear understanding of current research efforts, knowledge/research gaps, and funding patterns as scientific evidence is crucial to systematically accelerate the pace of research discovery in rare diseases, which is an overarching goal of this study.
To semantically represent NIH funding data for rare diseases and advance its use of effectively promoting rare disease research, we identified NIH funded projects for rare diseases by mapping GARD diseases to the project based on project titles; subsequently we presented and managed those identified projects in a knowledge graph using Neo4j software, hosted at NCATS, based on a pre-defined data model that captures semantics among the data. With this developed knowledge graph, we were able to perform several case studies to demonstrate scientific evidence generation for supporting rare disease research discovery.
Of 5001 rare diseases belonging to 32 distinct disease categories, we identified 1294 diseases that are mapped to 45,647 distinct, NIH-funded projects obtained from the NIH ExPORTER by implementing semantic annotation of project titles. To capture semantic relationships presenting amongst mapped research funding data, we defined a data model comprised of seven primary classes and corresponding object and data properties. A Neo4j knowledge graph based on this predefined data model has been developed, and we performed multiple case studies over this knowledge graph to demonstrate its use in directing and promoting rare disease research.
We developed an integrative knowledge graph with rare disease funding data and demonstrated its use as a source from where we can effectively identify and generate scientific evidence to support rare disease research. With the success of this preliminary study, we plan to implement advanced computational approaches for analyzing more funding related data, e.g., project abstracts and PubMed article abstracts, and linking to other types of biomedical data to perform more sophisticated research gap analysis and identify opportunities for future research in rare diseases.
A rare disease is defined as any disease that affects fewer than 200,000 individuals in the United States. There are an estimated 25–30 million Americans that are affected by one of approximately 7000 different rare diseases, most of which are poorly understood with unclear underlying biological mechanisms. This knowledge gap leads to challenges for patients, clinicians, and investigators. Patients affected by a rare disease experience delays in diagnosis, as well as a lack of available treatments, clinicians often have limited clinical knowledge and experience impedes their clinical decision making, and investigators struggle with limited patient data and sparse funding for research across most rare diseases [ 1 ]. To help address these challenges, we proposed a detailed analysis of research funding data to (1) enhance understanding of the current funding situation and potential funding opportunities in rare diseases, and (2) identify gaps among current research activities in rare diseases that may be primed for new research.
Compared to common diseases that are more highly prevalent in the population, such as depression or heart disease, research funding is often scarce for rare diseases, in part due to the relatively small number of people affected, and lower prioritization of funding based on the perceived burden of the disease [ 2 ]. In fiscal year 2019, the National Institutes of Health (NIH) were appropriated $39 billion [ 3 ], of which only $38 million (0.1%) was awarded to study a wide range of rare diseases [ 4 ]. Similarly, in the UK, less than 1% of the annual funding budget from three out of the top four UK funders were directed towards rare disease research [ 5 ]. A lack of funding results in less research aiming to understand disease etiology, identify biomarkers for disease diagnosis, develop of novel medications and associated clinical trials, and ultimately, absent treatment options. To solicit and attract more funding for innovative research in rare diseases, there is an urgent need to better understand and assess the current funding situation/trend and address gaps found to persist from retrospectively tracing funding history.
To review the funding landscape, Stehr et al. examined funding circumstances for Batten Disease, a group of rare nervous system disorders, by extracting funding information from publications. Interestingly they discovered 193 funding agencies had supported Batten Disease research to date, which might encourage researchers to continue their pursuits and expand their studies, moving key findings from discovery to application phases [ 6 ]. Franceschi et al. led a study to characterize recent NIH funding for diagnostic radiology departments at US medical schools [ 7 ]. To inform decisions on research direction, Ma et al. examined over 43,000 scientific projects funded over the past three decades and established collaboration networks that revealed major ramifications on future research strategy and government policy [ 8 ]. Packalen et al. performed an analysis on a comprehensive corpus of published biomedical research articles, and found that edge science framed with novel basic science ideas is more often funded by the NIH than less novel science [ 9 ]. These studies either limited their review of the funding landscape to a few specific diseases or diagnostic protocols, or assessed research direction globally. In our study, we aimed to develop an approach to systematically overview funding trend across different disease categories or individual diseases, and identify research gaps globally or locally (i.e., for individual disease category or disease) to further support rare disease research.
To overcome the challenge of handling large amounts of data accumulated given scientific advancements, knowledge graphs have attracted a lot of interest in the biomedical domain, as they can be leveraged to semantically represent relationships among large-scale data [ 10 ]. Ding, et al. constructed a PubMed knowledge graph (PKG) by extracting bio-entities from 29 million PubMed abstracts with integrated funding data through the NIH ExPORTER, in order to measure scholarly impact, knowledge usage and transfer, and profile authors and organizations based on their connections with bio-entities [ 11 ]. In our previous work, we developed an integrative knowledge graph in Neo4j [ 12 ], named NCATS GARD Knowledge Graph (NGKG) that contained a large volume of biomedical and research data pertinent to rare diseases [ 13 ]. Inspired by these published studies, we proposed to aggregate and represent funding data for rare diseases in a semantic manner as a knowledge graph. While the PKG incorporates publications and funding to gain insights and approaches to connect researchers with common research interests, our primary goal is to not only assess the funding landscape of rare diseases, but also identify research status and knowledge gaps, which can be applied to promote novel research for rare diseases.
Rare disease resources
In this study, we incorporated rare disease information from the Genetic and Rare Diseases (GARD) [ 14 ] and Monarch Disease Ontology (MONDO) [ 15 ]. GARD is a public health information center managed by the Office of Rare Diseases Research (ORDR) within the National Center for Advancing Translational Sciences (NCATS), and retains curated disease information for about 7000 rare diseases. MONDO is a semi-automatically constructed ontology that merges multiple disease resources to yield a single coherent merged ontology [ 15 ]. We accessed GARD and MONDO from our previously developed NCATS GARD Knowledge Graph (NGKG) in Neo4j [ 13 , 16 ].
NIH funding resource
Research Portfolio Online Reporting Tools Expenditures and Results (RePORTER) is a key component of the NIH’s “open government” initiative to provide more transparency into NIH activities, including information on NIH expenditures and the results of NIH supported research [ 17 ]. ExPORTER provides bulk administrative data found in RePORTER to the public for detailed analyses or to load into their own data systems [ 18 ]. ExPORTER provides downloadable versions of the data accessed through the RePORTER interface, and includes information about projects, publications, patents, and clinical studies. In this study, we downloaded and cleaned the funded projects and associated publications from ExPORTER, and stored the data in a MySQL database, from where we obtained data for the analysis described in this study.
Rare disease data preparation
We extracted 6305 GARD rare diseases from the NGKG in Neo4j [ 13 , 16 ]. In order to adopt disease categories from MONDO to organize these extracted rare diseases, we only included 5001 GARD rare diseases that have one-to-one exact mapping to MONDO based on one MONDO property, “MONDO:equivalentTo”. As a proof-of-concept with minimized manual validation, we did not include one-to-many, many-to-many or any other partial mappings between GARD and MONDO concepts, which will be included in a future study.
To enable review and analysis of research funding by disease categories, we mapped GARD diseases to MONDO disease categories. MONDO contains three main branches in its disease classification tree, namely, “Disease Characteristic”, “Disease or Disorder” and “Disease Susceptibility”. In this study, we focused on the branch of “Disease or Disorder” (MONDO: 0000001) in MONDO obo file [ 19 ], from where we extracted 32 root disease categories, including congenital abnormality, acute disease, disorder involving pain, serpinopathy, psychiatric disorder, visceral myopathy, and post-infectious disorder, etc. (A complete list of 32 root disease categories can be found in Additional file 1 ) We mapped those 5001 GARD diseases to the 32 root categories accordingly by iteratively searching the MONDO disease hieratical tree. It is worthy to note that most GARD diseases map to more than one MONDO disease categories.
NIH funded project identification for rare diseases
NIH ExPORTER provides detailed information about funded projects, including project titles and project abstracts in free text. We assumed that if the disease name is mentioned in the project title, it is likely that the project was proposed to conduct research investigation on this disease. Hence, we proposed to map GARD disease names to project titles, to identify a list of funded projects for each individual GARD disease via two steps as described below. Before mapping, we excluded projects with invalid project titles, such as, “13.358”, “CFDA NO. 13-299”.
Mapping based on name match
We executed ‘LIKE’ SQL operator as exact string match to identify projects with disease names mentioned in the project titles from the MySQL database, which stores the cleaned funding data. To avoid any mis-mappings, we applied not only full disease names but also alternative disease names in the ‘LIKE’ operator. Since abbreviations occurring in disease names and/or synonyms might cause incorrect mappings, disease names and synonyms with less than 4 characters were excluded for mapping.
Mapping based on semantic annotation.
As a complementary step, we semantically annotated project titles in free text by using MetaMap annotator [ 20 ], and then mapped GARD diseases to those generated annotations. MetaMap annotator produces a list of annotations in the Unified Medical Language System (UMLS), including UMLS semantic types, its preferred names, etc. Figure 1 shows a snapshot of annotation results generated for a project entitled “The Natural History of Mucolipidosis Type IV”. To avoid any incorrect mappings, we excluded those annotated concepts with less than 4 characters for further analysis, for example: “RNS (UMLS: C1850106)” as one annotation for the project title of “HAMPTON INSTITUTE'S CONTINUING EDUCATION PROGRAM FOR RNS”.

Annotation results generated by MetaMap (The fields in bold were extracted and applied to map to GARD diseases, “C0238386” is UMLS identifier, “Mucolipidosis Type IV” is UMLS preferred name, “[dsyn]” is one UMLS semantic type, “Disease or Syndrome”)
In order to establish the mappings between funded projects and GARD diseases, we mapped annotations generated from each project title to GARD diseases based on UMLS mappings since MetaMap output is in the UMLS, as shown in Fig. 1 . We retrieved mappings between GARD diseases and the UMLS via two steps. First, we obtained the UMLS mappings that were curated by GARD. Next, we obtained the UMLS mapping from MONDO for the GARD disease if the GARD disease concept was exactly matched to the MONDO concept and this MONDO concept had an external mapping to the UMLS. For instance, Bloom syndrome (GARD:0000915) is exactly matched to one MONDO concept of Bloom syndrome (MONDO:0008876) that is mapped to one UMLS concept (UMLS:C0005859). Thus, with UMLS:C0005859, we were able to map this GARD disease to one project entitled “BLOOM'S SYNDROME--DNA LIGASE AND IMMUNODEFICIENCY” with one annotation of Bloom Syndrome (UMLS:C0005859).
Once we established the connections between GARD diseases and funded projects through the above steps, we designed a data model to semantically capture and represent different types of data extracted from those funded projects and their associated data, such as, publications or principal investigators.
Primary class definition
We defined seven primary classes, namely, Disease Category, Disease, Funded Project, Funding Agent, Principal Investigator, Publication, and Journal. These classes capture a full spectrum of information present in NIH funding data and enable linkages to other different types of data for directing more sophisticated research on rare diseases, which will be described in the “ Discussion ” section.
Object property definition
To capture semantic relationships among those primary classes, we defined object properties as shown in Table 1 .
Data property definition
We defined a list of data properties shown in Table 2 to link data values for each individual concept.
NIH funding knowledge graph
Based on the data model we described above, we loaded the mapped funding data to a knowledge graph hosted in Neo4j. To be specific, different types of data have been loaded and represented with those seven primary classes as nodes accordingly; object properties were applied to establish semantic connections between different nodes as edges, and data properties were attached to corresponding nodes as node properties. The knowledge graph is publicly assessable without login requirement at http://grants4rd.ncats.io:7474/browser/ .
Results of rare disease data preparation
A total of 5001 unique GARD rare diseases were categorized based on the MONDO disease classes. Table 3 shows the categorization results. Only 799 GARD diseases belonged to a single MONDO category, while most GARD diseases were mapped to multiple MONDO disease categories. For example, GARD:0006735 (Hypophosphatemic rickets) is mapped to three different MONDO disease categories: MONDO:0003847 (Mendelian disease), MONDO:0003900 (connective tissue disease), and MONDO:0021199 (disease by anatomical system). There are 52 GARD diseases that were not grouped into any of MONDO disease categories, because they were either mapped to obsolete MONDO diseases or another two MONDO disease category branches, “Disease Characteristic” and “Disease Susceptibility”, which were excluded from this study.
Results of NIH funding data mapping
Results of nih funding data retrieval.
We downloaded the funding data with funding year spanning from 1985 to 2019 from NIH ExPORTER [ 18 ]. A total of 2,457,303 distinct applications with 654,347 unique project titles, 886,895 unique project abstracts, and 2,555,300 publications are cleaned and stored in a MySQL database.
Results of NIH funding data mapped to GARD
Two types of mapping results have been generated via exact name (i.e., String) match and MetaMap annotations.
Disease name mapping results
Both GARD disease names and synonyms have been applied to map project titles. 1104 GARD diseases were mapped to 21,027 project titles, which correspond to 63,692 NIH funded applications. Since one project can be funded for multiple years as multiple applications with the same project title, the number of mapped applications is larger than the number of project titles.
MetaMap annotation results
652,975 unique project titles were annotated by the MetaMap annotator and 5,039,735 annotations were generated. To map GARD diseases to those annotations based on UMLS mappings, we first retrieved UMLS mappings for GARD diseases from GARD and MONDO. Specifically, 3468 GARD diseases with UMLS mappings were extracted from GARD, and out of 15,629 MONDO concepts with UMLS mappings, 3980 MONDO entries were mapped to 4032 GARD diseases and corresponding UMLS mappings, which were assigned to those GARD diseases. Together, 1146 GARD diseases were mapped to 13,695 project titles via UMLS mappings.
By merging mapping results from the above two steps, 1294 GARD diseases were successfully mapped to 72,577 funded applications, which corresponds to 45,647 distinct projects.
Statistical results of NIH rare disease funding data in the Neo4j knowledge graph
Summarized statistical results of individual concepts belonging to each primary class are shown in Fig. 2 .
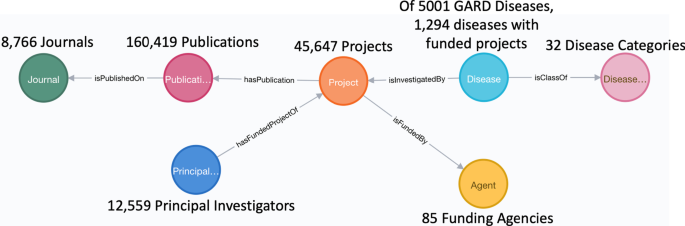
Statistics of rare disease funding Neo4j knowledge graph
Case studies
To demonstrate the use of this integrated knowledge graph with rare disease funding data, we performed two types of case studies, (1) funding landscape assessments for an overview of the current funding landscape; (2) evidence-based research opportunity identification for supporting research in rare diseases. Cypher Queries have been composed and executed to perform case studies described in this section, and more details about those Cypher Queries are included in the Additional file 2 .
Current funding landscape
To review NIH funding scenario in rare diseases, we composed several queries using Cypher, which is Neo4j’s graph query language that allows users to store and retrieve data from the graph database [ 21 ], to search against funding data in our Neo4j. Cypher Query 1 was constructed to access funding circumstances by disease categories and the result is listed in Table 4 . Since one disease might be grouped into multiple disease categories, duplicates occurred when the number of GARD diseases, number of projects and total cost by each category were summed. Regardless, the numbers listed in Table 4 consistently reflect funding priorities with the consideration of disease burden applied by NIH. In addition, we retrieved funding amounts in the last twenty years for the top five most funded disease category, which is shown in Fig. 3 . It is worthy to note that no funding cost data is available before the year of 2000 in our downloaded data. From Fig. 3 , besides infectious diseases, big jumps were present due to the Ebola and Zika outbreaks that occurred during that time frame, the increasing funding trends are observed for other categories.
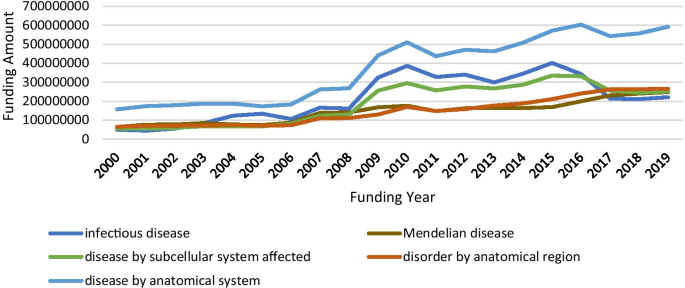
Funding trend by year for the top five most funded disease categories
Cypher Query 1
MATCH p = (n:Project)<-[:isInvestigatedBy]-(d:Disease)-[:isClassOf]-(c:DiseaseCategory) RETURN c.name AS Disease_Category, COUNT(DISTINCT d.gard_id) AS NumOfGARD, COUNT(n.application_id) AS NumOfProjects, SUM(TOINTEGER(n.total_cost)) AS Total_Funding_Amount ORDER BY Total_Funding_Amount DESC
We also evaluated the top 10 funded individual diseases (via Cypher Query 2). The top four funded diseases shown in Table 5 are infectious diseases. Poliomyelitis is considered a global public health emergency and the U.S. is a partner of the Global Polio Eradication Initiative [ 22 ]. Particularly, the CDC and the U.S. Agency for International Development (USAID) work to eradicate polio and have signed onto the Polio Endgame Strategy 2019–2023 [ 23 ]. About half of the world population is at risk for Malaria and the U.S. is the largest donor to the Global Fund to Fight AIDS, Tuberculosis, and Malaria (Global Fund) [ 24 ]. A large amount of research funds are being distributed on research for effective vaccines for Anthrax and Measles. In comparison to other rare diseases, the rest of diseases listed in Table 5 , such as sickle cell disease with about 100,000 affected people in the US, both of cystic fibrosis and Huntington disease with more than 30,000 affected people in the US, are diseases that are closer to finding a cure or effective treatment, in part because they receive more research funding.
Cyper Query 2
MATCH p = (d:Disease)-[:isInvestigatedBy]-(n:Project) RETURN d.gard_id AS GARD_ID, d.name AS GARD_Name, SUM(toInteger(n.total_cost)) AS Total_Funding_Amount ORDER BY total_cost DESC LIMIT 10
Evidence-based rare disease research discovery
We aggregated funded projects and their publications into an integrated knowledge graph in Neo4j, which offers opportunities to programmatically support new research for rare diseases.
Research landscape assessment
Funded projects along with their publications show snapshots of their research goals and outcomes, which provides an opportunity to systematically assess the current research status and gaps, and consequently direct future areas for investigation. We enumerated two types of assessment for Neuronal ceroid lipofuscinosis (GARD: 0010739) and Duchenne Muscular Dystrophy (GARD:0007922) as examples.
Research status assessment for neuronal ceroid lipofuscinosis (NCL)
NCLs are classified by their causal gene of CLN (ceroid lipofuscinosis, neuronal), which is given a different number designation as its subtype. Signs and symptoms range in severity and progress at different rates given different gene mutations. The disorders generally include a combination of vision loss, epilepsy, and dementia. Some forms of the NCLs are: CLN1 disease, infantile onset; CLN2 disease, later-onset and so on [ 25 ]. Scientific investigations have been performed for each subtype of NCL (via Cypher Query 3). By manually examining the funded projects and their published studies for NCL, we grouped them based on their studied NCL forms (Fig. 4 ). Noticeably, these funded projects aim to better understand the molecular mechanism of NCL and discover therapeutic solutions. In particular, several subtypes (CLN2, CLN3 and CLN6) of NCL are more extensively studied than others. There is an Food and Drug Administration (FDA) approved enzyme replacement therapy for CLN2 disease (TTP1 deficiency) called cerliponase alfa (Brineura®) that has been shown to slow or halt the progression of symptoms [ 26 ]. With the exception of CLN2, which is highlighted as “CLN2” in Fig. 4 , there are no approved treatments that can slow or stop disease progression for other forms of NCL disorder. As NCL affects the brain and nervous system, treatments must reach the brain to be effective, but getting the proper enzyme to cross the blood brain barrier can be difficult. For this reason, enzyme replacement therapy can only be used in NCL forms where the affected enzyme is soluble. This includes the subtypes known as CLN1, CLN2, CLN5 and CLN10 [ 27 ]. Thus, learning from the success of Brineura for CLN2 enhances the necessary scientific evidence and understanding of what existing research has been performed in other forms of NCL, and may better inform the direction of further research investigation on NCL.
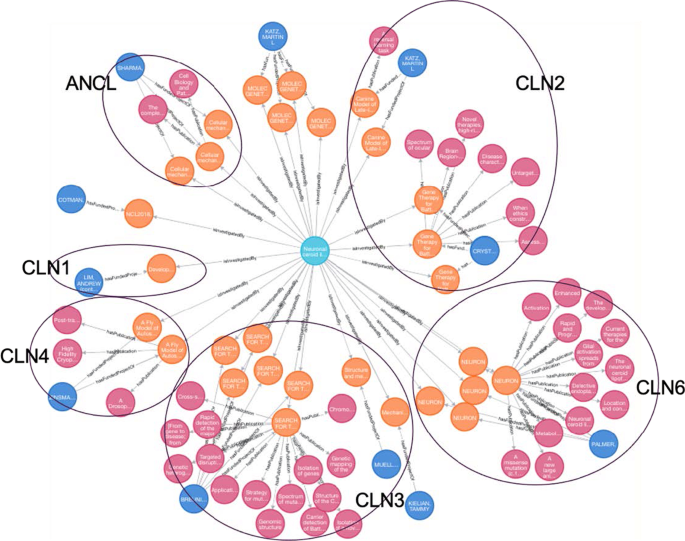
Research landscape of Neuronal ceroid lipofuscinosis (Light blue nodes denote diseases, orange nodes denote funded projects, magenta nodes denote publications, Dark blue nodes denote principal investigators. CLN1, CLN2, CLN3, CLN4, CLN6 and ACLN are different forms of NCL)
Cypher Query 3
MATCH p = (d:Disease)-[:isInvestigatedBy]->(n:Project)<-[:hasFundedProjectOf]-(m:PrincipalInvestigator) WHERE d.gard_id = 'GARD:0010739' RETURN p
Research study clustering for Duchenne Muscular Dystrophy
Publications introduce novel approaches and/or findings that were proposed and generated from the funded projects. For example, a project entitled “LOCALIZATION OF X-LINKED HYPOPHOSPHATEMIC RICKETS GENE” (APPLICATION ID = “3087091”), has reported two publications, “Mutational analysis and genotype-phenotype correlation of the PHEX gene in X-linked hypophosphatemic rickets.” (PMID: 11502829) and “Mutational analysis of the PEX gene in patients with X-linked hypophosphatemic rickets.” (PMID: 9106524) These two publications specifically reported their investigation on genes for X-linked hypophosphatemic rickets. For a given disease, we clustered research studies by a project or a list of projects with similar research topics, which allows tracking research trajectory, identifying research gaps, and preparing necessary training data for future study. In this case study, we executed Cypher Query 4 to cluster publications associated with the funded projects for Duchenne Muscular Dystrophy (GARD:0006291), a lethal muscle wasting disease caused by the lack of dystrophin, which eventually leads to apoptosis of muscle cells and impaired muscle contractility. Four projects, along with their publications, are shown in Fig. 5 . Based on the project titles shown in gray boxes in Fig. 5 , we deduced that their objectives are all tied to breaching the major barriers to successful therapeutic solutions for Duchenne Muscular Dystrophy respectively via iPSCs, Cas9 and Cx43.
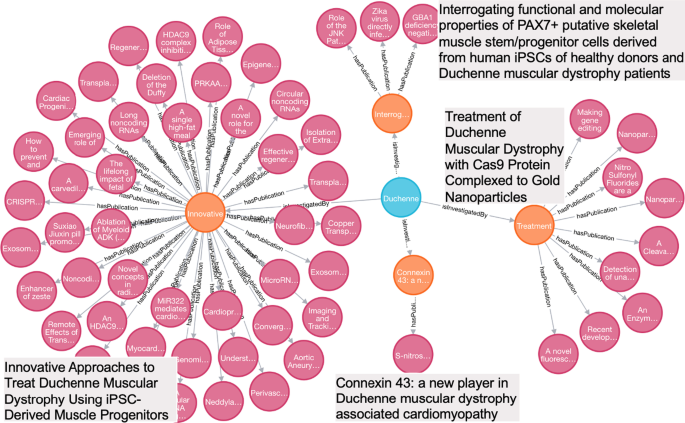
Publications clustered by funded projects for Duchenne Muscular Dystrophy (Blue nodes denote diseases, orange nodes denote funded projects, magenta nodes denote publications. Project titles are shown in the gray boxes)
Cypher Query 4
MATCH p =(d:Disease)-[:isInvestigatedBy]-(n:Project)-[:hasPublication]-(m:Publication) WHERE d.gard_id = 'GARD:0006291' RETURN p
Identify new research opportunities
In this case study, we proposed to gain research insights systematically by analyzing project titles. For instance, given one project entitled “USE OF LEUPROLIDE ACETATE FOR TREATMENT OF PRECOCIOUS PUBERTY”, we know this project was studying leuprolide acetate (Drug) for precocious puberty [ 28 ]. With the disease annotations generated for project titles (described in the “ Identify new research opportunities ” section), we were able to identify potential disease associations via projects, and furthermore to identify new avenues for research. Fig. 6 demonstrates an example of looking for potentially relevant diseases to Measles (GARD:0003434) via Cypher Query 5. Obviously multiple rare infectious diseases, including Poliomyelitis, Rubella, Congenital rubella, and Malaria are grouped together, and interestingly Measles and Glioblastoma is linked via one project entitled “Measles Virotherapy for Glioblastoma Multiforme”, highlighted in the red circle shown in Fig. 6 . According to one recently published review paper written by Zhang et al. [ 29 ], “advances and potential pitfalls of oncolytic viruses expressing immunomodulatory transgene therapy for malignant gliomas”, the authors emphasized that the therapeutic efficacy of oncolytic viruses alone is limited, which might motivate novel research to better understand and assess the therapeutic efficacy. Five related publications produced by this project provide additional information to direct future investigation.

Identifying possible research opportunities for Measles via funded projects (Orange nodes denote funded projects, blue nodes denote rare diseases, magenta nodes denote publications)
Cypher Query 5
MATCH p =(d:Disease)-[:isInvestigatedBy]-(n:Project)-[:isInvestigatedBy]-(d1:Disease) WHERE d.gard_id = 'GARD:0003434' RETURN p
In this study, we extracted NIH funding data for rare diseases and semantically represented it as a knowledge graph, which provides an effective way to not only assess the funding landscape in rare diseases, and systematically identify new research gaps/opportunities in rare diseases, but also share identified scientific findings/evidence with funding agencies for recommending their research fund distribution in rare diseases.
Scientific observations from data preparation
Given various representations of disease names applied in the project titles, to accurately map diseases to projects via project titles and avoid any mis-mapping, we applied a complementary mapping strategy using string match and semantic annotation. For instance, one project entitled “Discovering Novel Treatments for Batten Disease” could only be mapped to “Batten Disease (GARD:0010739)” via string match based on the disease name, instead of semantic annotation. The MetaMap annotations generated from the project title is “Juvenile Neuronal Ceroid Lipofuscinosis (UMLS: C0751383)”, which is different from the UMLS mapping (UMLS:C0027877) associated with this GARD disease. Another project entitled “TOXIC METALS, MEMBRANE SIGNALING, AND CELL GROWTH” is mapped to “Heavy metal poisoning (GARD:0006577)” via semantic annotation instead of string match, since the MetaMap annotator generates one annotation of “toxic metals (UMLS: C0274869)”, which is mapped to the UMLS associated with that GARD disease. Clearly, even though we applied this complementary strategy to establish mappings between projects and diseases, we may miss relevant projects with the disease mentioned in the project abstract, instead of in the project title. To avoid any missing data, several additional steps have been proposed and described in the “ Future work ” section.
Scientific insights derived from the case studies
We performed two types of case studies, funding landscape and research landscape analysis respectively to demonstrate the use of our funding knowledge graph for evidence-based rare disease research discovery.
Funding landscape assessment
Accessing funding data allows researchers to examine funding situations and funding opportunities in rare diseases. Our case studies illustrate that the funding trend consistently reflects the disease burden to the public health, e.g., infectious diseases received the most amount of funding support over years. A big jump regarding the total funding amount for infectious diseases between 2013 and 2017, coincides with the Ebola and Zika outbreaks that occurred during that timeframe. Another inspiring message derived from our analysis is that the amount of funding for supporting rare disease research has been increasing yearly, which might encourage and motivate more scientists to devote more of their research to rare diseases.
Scientific evidence-based rare disease research discovery
Analysis of funding data not only provides a systematic way to outline a complete research spectrum for an interested disease (category), but also generates scientific evidence supporting rare disease research discovery. For instance, the case study of Neuronal Ceroid Lipofuscinosis (NCL) illustrates that several different types of NCL have been investigated intensively and one therapeutic success on one type of NCL, namely, CLN2, which will enable us to dive into it and derive insight and knowledge about the status and gaps of research on NCL in further investigation. In addition, we demonstrated the power of aligning funded projects with their publications to direct research. As shown in the case study of Duchenne Muscular Dystrophy, it becomes feasible to cluster research papers based on a funded project or a list of projects with a similar research topic, which enables researchers to systematically track research pathway and programmatically prepare training data for future study. In this study, publication clusters were solely based on single projects; we propose to cluster publications with multiple projects with similar research topics by implementing Natural Language Processing (NLP) algorithms to analyze project abstracts.
Limitations and future work
By reviewing the MONDO categories applied to organize rare diseases, some categories, for instance, “Mendelian disease”, “disease by anatomical system”, are broad categories consisted of many individual diseases; In addition, as shown in Table 3 , most rare diseases are grouped into multiple MONDO categories. We propose to extend our disease category mapping to a higher granularity level as the next step, to precisely reflect funding distribution and assess research landscape by disease categories.
In this study, mappings between rare diseases and funded projects were established based on project titles. To avoid any mis-mappings with rare diseases that are mentioned in the project abstracts instead of project titles, we propose to expand our analysis with project abstracts. In addition, analyzing funding data from other resources, such as “funding” or “Acknowledgements” sections included in the publications to introduce the funding sources supporting their research, is another proposed extension task. Although the aforementioned extensions have been proposed as future work by aligning with our current project plan, we anticipated that we will still miss some data, such as, (1) projects without project titles and/or abstracts, (2) non-US government funded projects; (3) projects with only a single funding source (not all) was acknowledged in the publications. These are beyond the scope of this project and will be planned in the future study.
Insights and lessons drawn from the case studies were heavily relied on manual interpretation in this preliminary study, and we propose to transform currently manual processes into automated processes by implementing NLP and advanced machine learning techniques. Given the nature of rare diseases, limited data and knowledge about those diseases compared to common diseases, collaborative efforts seem more important and critical. We propose to discover methods to better promote research collaboration by connecting investigators with the requisite expertise and shared research interest from our funding knowledge graph. To that end, scientists can work collaboratively to pool patients, data, experience and resources together to support more innovative research in rare diseases. We are also interested in investigating the relationship between medical cost using information from datasets such as the AHRQ’s Healthcare Cost and Utilization Project (HCUP) [ 30 ] and research cost (i.e., NIH funding) in rare diseases, with the hypothesis that higher research funding should lead to improvements in earlier diagnosis of a rare disease and lower medical costs. Identifying overlaps and discrepancies between research status and medical situation will guide both further investigation and better inform decision makers on how to stimulate and advance research across more rare diseases.
In this preliminary study, we developed an integrative knowledge graph to semantically represent NIH research funding data for rare diseases and successfully demonstrated its use of directing and promoting scientific evidence based rare disease research discovery. With the success and lessons learned from this study, we propose multiple improvements/extensions as the next step, to fully utilize funding data to accelerate the pace of rare disease research.
Availability of data and materials
The data applied in this study can be accessed at http://grants4rd.ncats.io:7474/browser/ .
Abbreviations
Genetic and Rare Disease Information Center
Mondo Disease Ontology
NCATS GARD Knowledge Graph
Unified Medical Language System
Open Biological and Biomedical Ontologies
Natural Language Processing
Stoller JK. The challenge of rare diseases. Chest. 2018;153(6):1309–14.
Article Google Scholar
Gross CP, Anderson GF, Powe NR. The relation between funding by the National Institutes of Health and the burden of disease. N Engl J Med. 1999;340(24):1881–7.
Article CAS Google Scholar
We have a budget for FY 2019! https://www.nia.nih.gov/research/blog/2018/10/we-have-budget-fy-2019#:~:text=The%20total%20NIH%20appropriation%20for,Alzheimer's%20and%20related%20dementias%20research .
NIH funding bolsters rare diseases research collaborations. https://www.nih.gov/news-events/news-releases/nih-funding-bolsters-rare-diseases-research-collaborations .
Why is rare disease funding rare? https://pharmaphorum.com/views-and-analysis/why-is-rare-disease-funding-rare/ .
Stehr F, Forkel M. Funding resources for rare disease research. Biochim Biophys Acta: BBA Mol Basis Dis. 2013;1832(11):1910–2.
Franceschi AM, Rosenkrantz AB. Patterns of recent National Institutes of Health (NIH) funding to diagnostic radiology departments: analysis using the NIH RePORTER system. Acad Radiol. 2017;24(9):1162–8.
Ma A, Mondragón RJ, Latora V. Anatomy of funded research in science. Proc Natl Acad Sci. 2015;112(48):14760–5.
Packalen M, Bhattacharya J. NIH funding and the pursuit of edge science. Proc Natl Acad Sci. 2020;117(22):12011–6.
Yoon B-H, Kim S-K, Kim S-Y. Use of graph database for the integration of heterogeneous biological data. Genomics Inform. 2017;15(1):19.
Xu J, Kim S, Song M, Jeong M, Kim D, Kang J, et al. Building a PubMed knowledge graph. Sci Data. 2020;7(1):1–15.
Lal M. Neo4j graph data modeling. Birmingham: Packt Publishing Ltd; 2015.
Google Scholar
Zhu Q, Nguyen D-T, Grishagin I, Southall N, Sid E, Pariser A. An integrative knowledge graph for rare diseases, derived from the Genetic and Rare Diseases Information Center (GARD). J Biomed Semant. 2020;11(1):1–13.
The Genetic and Rare Diseases Information Center (GARD). https://rarediseases.info.nih.gov/ .
Mondo Disease Ontology. https://mondo.monarchinitiative.org/ .
NCATS Integrated Disease Ontologies. https://disease.ncats.io/browser/ .
NIH RePORT. https://report.nih.gov/ .
NIH ExPORTER. https://exporter.nih.gov/default.aspx .
Mondo Disease Ontology. http://www.obofoundry.org/ontology/mondo.html .
MetaMap. https://metamap.nlm.nih.gov/ .
Cypher Query Language - Developer Guides - Neo4j. https://neo4j.com/developer/cypher/ .
Global Polio Eradication Initiative (GPEI). https://polioeradication.org/ .
Polio Endgame Strategy. https://polioeradication.org/who-we-are/polio-endgame-strategy-2019-2023/#:~:text=The%20GPEI%20Polio%20Endgame%20Strategy,world%20free%20of%20all%20polioviruses.&text=The%20GPEI%20Polio%20Endgame%20Strategy%202019%2D2023%20was%20developed%20in,stakeholders%20from%20around%20the%20world .
TheGlobalFund. https://www.theglobalfund.org/en/ .
Batten Disease Fact Sheet. https://www.ninds.nih.gov/Disorders/Patient-Caregiver-Education/Fact-Sheets/Batten-Disease-Fact-Sheet .
Markham A. Cerliponase alfa: first global approval. Drugs. 2017;77(11):1247–9.
Enzyme Replacement Therapy. https://battendiseasenews.com/enzyme-replacement-therapy/ .
FDA approves twice-yearly leuprolide injection for central precocious puberty. https://www.healio.com/news/endocrinology/20200504/fda-approves-twiceyearly-leuprolide-injection-for-central-precocious-puberty#:~:text=A%20leuprolide%20acetate%20suspension%20to,in%20a%20polymeric%20gel%20form .
Zhang Q, Liu F. Advances and potential pitfalls of oncolytic viruses expressing immunomodulatory transgene therapy for malignant gliomas. Cell Death Dis. 2020;11(6):1–11.
AHRQ’s Healthcare Cost and Utilization Project (HCUP). https://www.ahrq.gov/data/hcup/index.html .
Download references
Open Access funding provided by the National Institutes of Health (NIH). This research was supported by the Intramural and Extramural research program of the NCATS, NIH.
Author information
Authors and affiliations.
Division of Pre-Clinical Innovation, National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (NIH), Rockville, MD, 20850, USA
Qian Zhu, Ðắc-Trung Nguyễn, Timothy Sheils & Ewy A. Mathé
ICF International Inc, Rockville, MD, USA
Gioconda Alyea
Office of Rare Diseases Research, National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (NIH), Bethesda, MD, 20892, USA
Eric Sid, Yanji Xu, James Dickens & Anne Pariser
You can also search for this author in PubMed Google Scholar
Contributions
The work was conceived by QZ, who designed and performed data analysis and drafted the manuscript; TN helped on Neo4j development; GA helped on case studies with funding landscape assessment; ES, EAM helped on case studies with research landscape assessment and manuscript edition; TS, YX, JD and AP participated in the project discussion and edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Correspondence to Qian Zhu .
Ethics declarations
Ethics approval and consent to participate.
Not applicable.
Consent for publication
Competing interests.
None declared.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
. 32 root disease categories from MONDO.
Additional file 2
. Cypher queries from the case studies.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Zhu, Q., Nguyễn, ÐT., Sheils, T. et al. Scientific evidence based rare disease research discovery with research funding data in knowledge graph. Orphanet J Rare Dis 16 , 483 (2021). https://doi.org/10.1186/s13023-021-02120-9
Download citation
Received : 13 May 2021
Accepted : 06 November 2021
Published : 18 November 2021
DOI : https://doi.org/10.1186/s13023-021-02120-9
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
Orphanet Journal of Rare Diseases
ISSN: 1750-1172
- Submission enquiries: Access here and click Contact Us
- General enquiries: [email protected]
- U.S. Department of Health & Human Services

- Virtual Tour
- Staff Directory
- En Español
You are here
Nih research matters.
December 22, 2021
2021 Research Highlights — Promising Medical Findings
Results with potential for enhancing human health.
With NIH support, scientists across the United States and around the world conduct wide-ranging research to discover ways to enhance health, lengthen life, and reduce illness and disability. Groundbreaking NIH-funded research often receives top scientific honors. In 2021, these honors included Nobel Prizes to five NIH-supported scientists . Here’s just a small sample of the NIH-supported research accomplishments in 2021.
Printer-friendly version of full 2021 NIH Research Highlights
20210615-covid.jpg
Advancing COVID-19 treatment and prevention
Amid the sustained pandemic, researchers continued to develop new drugs and vaccines for COVID-19. They found oral drugs that could inhibit virus replication in hamsters and shut down a key enzyme that the virus needs to replicate. Both drugs are currently in clinical trials. Another drug effectively treated both SARS-CoV-2 and RSV, another serious respiratory virus, in animals. Other researchers used an airway-on-a-chip to screen approved drugs for use against COVID-19. These studies identified oral drugs that could be administered outside of clinical settings. Such drugs could become powerful tools for fighting the ongoing pandemic. Also in development are an intranasal vaccine , which could help prevent virus transmission, and vaccines that can protect against a range of coronaviruses .
202211214-alz.jpg

Developments in Alzheimer’s disease research
One of the hallmarks of Alzheimer’s is an abnormal buildup of amyloid-beta protein. A study in mice suggests that antibody therapies targeting amyloid-beta protein could be more effective after enhancing the brain’s waste drainage system . In another study, irisin, an exercise-induced hormone, was found to improve cognitive performance in mice . New approaches also found two approved drugs (described below) with promise for treating AD. These findings point to potential strategies for treating Alzheimer’s. Meanwhile, researchers found that people who slept six hours or less per night in their 50s and 60s were more likely to develop dementia later in life, suggesting that inadequate sleep duration could increase dementia risk.
20211109-retinal.jpg
New uses for old drugs
Developing new drugs can be costly, and the odds of success can be slim. So, some researchers have turned to repurposing drugs that are already approved for other conditions. Scientists found that two FDA-approved drugs were associated with lower rates of Alzheimer’s disease. One is used for high blood pressure and swelling. The other is FDA-approved to treat erectile dysfunction and pulmonary hypertension. Meanwhile, the antidepressant fluoxetine was associated with reduced risk of age-related macular degeneration. Clinical trials will be needed to confirm these drugs’ effects.
20210713-heart.jpg
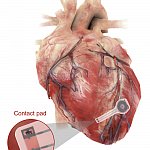
Making a wireless, biodegradable pacemaker
Pacemakers are a vital part of medical care for many people with heart rhythm disorders. Temporary pacemakers currently use wires connected to a power source outside the body. Researchers developed a temporary pacemaker that is powered wirelessly. It also breaks down harmlessly in the body after use. Studies showed that the device can generate enough power to pace a human heart without causing damage or inflammation.
20210330-crohns.jpg

Fungi may impair wound healing in Crohn’s disease
Inflammatory bowel disease develops when immune cells in the gut overreact to a perceived threat to the body. It’s thought that the microbiome plays a role in this process. Researchers found that a fungus called Debaryomyces hansenii impaired gut wound healing in mice and was also found in damaged gut tissue in people with Crohn’s disease, a type of inflammatory bowel disease. Blocking this microbe might encourage tissue repair in Crohn’s disease.
20210406-flu.jpg
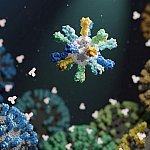
Nanoparticle-based flu vaccine
Influenza, or flu, kills an estimated 290,000-650,000 people each year worldwide. The flu virus changes, or mutates, quickly. A single vaccine that conferred protection against a wide variety of strains would provide a major boost to global health. Researchers developed a nanoparticle-based vaccine that protected against a broad range of flu virus strains in animals. The vaccine may prevent flu more effectively than current seasonal vaccines. Researchers are planning a Phase 1 clinical trial to test the vaccine in people.
20211002-lyme.jpg

A targeted antibiotic for treating Lyme disease
Lyme disease cases are becoming more frequent and widespread. Current treatment entails the use of broad-spectrum antibiotics. But these drugs can damage the patient’s gut microbiome and select for resistance in non-target bacteria. Researchers found that a neglected antibiotic called hygromycin A selectively kills the bacteria that cause Lyme disease. The antibiotic was able to treat Lyme disease in mice without disrupting the microbiome and could make an attractive therapeutic candidate.
20211102-back.jpg

Retraining the brain to treat chronic pain
More than 25 million people in the U.S. live with chronic pain. After a treatment called pain reprocessing therapy, two-thirds of people with mild or moderate chronic back pain for which no physical cause could be found were mostly or completely pain-free. The findings suggest that people can learn to reduce the brain activity causing some types of chronic pain that occur in the absence of injury or persist after healing.
2021 Research Highlights — Basic Research Insights >>
Connect with Us
- More Social Media from NIH
Centers for Research in Emerging Infectious Diseases (CREID)
The global network involves multidisciplinary investigations into how and where viruses and other pathogens can emerge from wildlife and spillover to cause disease in people. Research is led by 10 Centers and one Coordinating Center and will involve collaborations with peer institutions in the United States and 28 other countries.
Main Areas of Focus
Research projects will include surveillance studies to identify previously unknown viral causes of febrile illnesses in humans, the animal sources of viral or other disease-causing pathogens, and to determine what genetic or other changes make these pathogens capable of infecting humans. Other research by the CREID investigators will involve developing reagents and diagnostic assays to improve detection of emerging pathogens as well as studies aimed at detailing human immune responses to new or emerging infectious agents. Overall, the breadth of research projects to be carried out in the CREID network will allow for study of disease spillover in multiple phases of the process: where pathogens first emerge from an animal host; at the borders between wild and more populated areas, where and when rapid human-to-human transmission occurs; and, finally, how transmission is facilitated in urban areas, where epidemic spread can occur.
Primary awardees for the CREID network and regions of focus include:
- Donald Brambilla, Ph.D., RTI International, Research Triangle Park, North Carolina
- Tony Moody, M.D., Duke University School of Medicine, Durham, North Carolina CREID Coordinating Center
- Kristian Andersen, Ph.D., Scripps Research, La Jolla, California West African Emerging Infectious Disease Research Center West Africa
- Peter Daszak, Ph.D., EcoHealth Alliance, Inc., New York, New York Emerging Infectious Diseases Southeast Asia Research Collaboration Hub ( EID-SEARCH ) Southeast Asia
- Eva Harris, Ph.D., University of California, Berkeley American and Asian Centers for Arboviral Research and Enhanced Surveillance (A2CARES) Central and South America, South Asia
- Christine Johnson, VMD, Ph.D., University of California, Davis, School of Veterinary Medicine Epicenter for Emerging Infectious Disease Intelligence Central Africa and South America
- M. Kariuki Njenga, Ph.D., Washington State University, Pullman Center for Research in Emerging Infectious Diseases-East and Central Africa East and Central Africa
- Anavaj Sakuntabhai, Ph.D., Institut Pasteur, Paris, France West Africa and Southeast Asia
- Nikos Vasilakis, Ph.D., University of Texas Medical Branch, Galveston Central and South America
- Wesley C. Van Voorhis, M.D., Ph.D., University of Washington, Seattle South America, West and South Africa, Middle East, and Asia
- David Wang, Ph.D., Washington University School of Medicine, St. Louis, Missouri Asia, East Africa
- Scott C. Weaver, Ph.D., University of Texas Medical Branch, Galveston West Africa
Read more about this network: CREID Network
News and Announcements
Nov. 17, 2021: NIAID Supports Early-Career Researchers in Emerging Infectious Diseases
Aug. 27, 2020: NIAID Establishes Centers for Research in Emerging Infectious Diseases
Contact Information
Skip to site alert. Skip to content
Uhlemann Lab
Research projects, staphylococcus aureus.
The past decade has witnessed a dramatic increase in Staphylococcus aureus infections acquired from the community. Yet there remains a limited understanding of how these strains spread and subsequently become established and persist within communities. Our research is focused on identifying molecular mechanisms that allow epidemic S. aureus strains such as USA300 to successfully disseminate. This project uses a combined approach of whole-genome comparative sequencing of longitudinally collected samples, genetic manipulation, and functional studies on bacterial adhesion and survival. These molecular studies are informed by ongoing epidemiological studies on S. aureus transmission in the local community. This work has identified a potentially newly emerging S. aureus strain, ST398, which was previously only associated with close contacts to animals. Ongoing studies are aimed at elucidating the molecular mechanisms of its cross-species transfer and current animal-independent spread.
Klebsiella Pneumoniae Infections in Transplant Patients
Infections with carbapenem-resistant Enterobacteriaceae (CRE) have emerged as an urgent threat to healthcare, the “category of highest concern” in the 2013 CDC antimicrobial threat report. These multi-drug resistant infections are associated with high mortality. Recipients of solid organ transplantation, in particular liver transplant recipients are at increased risk for CRE infections. While intestinal colonization with CREs has been proposed as a potential risk factor for infections during CRE outbreaks, its actual contribution to infection remains incompletely understood. Moreover, there is a fundamental gap in knowledge on how these antibiotic resistant organisms transition from colonization to infection within affected hosts. We have established a cohort study of patients undergoing liver transplantation in which we aim to investigate the contribution of colonization to subsequent CRE infection. The long-term goal of this project is to elucidate at the bacterial genome level how CRE infections emerge and spread. Understanding these processes is critical to developing intervention and real-time clinical monitoring approaches to limit the impact of CRE infections at an individual and population level.
Microbiome in Infectious Diseases
During intestinal colonization, microorganisms closely interact with their surrounding bacterial communities and the immune system. Even single doses of antibiotics can profoundly affect the composition of the gut microbiota. The effects of organ transplantation on the microbiome are multifaceted, including the underlying disease, immunosuppressants, perioperative antibiotics, and the surgery itself. We are investigating how the intestinal microbiome modulates colonization with multi-drug resistant organisms in liver transplant recipients. Additional studies focus on the interaction of the intestinal and oral gut microbiome with the immune system in persons living with HIV.
Division of Infectious Diseases section navigation
Infectious diseases research.
The Division of Infectious Diseases engages in a broad spectrum of basic, clinical, translational, and epidemiologic sciences. The division garners more than $30 million in research funding per year.

Infectious Diseases faculty collaborate actively with other top-rated local and regional programs, including the Emory Vaccine Center , the Emory Department of Microbiology and Immunology , the Emory Rollins School of Public Health , and the Centers for Disease Control and Prevention (CDC) , in addition to numerous centers in the United States and abroad.
You can view ongoing trials in Infectious Diseases by visiting Emory Clinical Trials .
Research Programs
- Antibiotic Resistance Center
- Center for AIDS Research
- Kwazulu-Natal Drug Resistance Surveillance Study
- Emerging Infections Program
- Emory Clinical Trials Unit
- Emory T32 Training Program in Translational Research to End the HIV Epidemic
- Hope Clinic - Vaccine Research
- Prevention Epicenter of Emory and Collaborating Healthcare Facilities (PEACH)
- Ponce Clinical Research Site
- Infectious Diseases Clinical Research Collaborative
- Integrating HIV and heart health in South Africa (iHEART-SA) project
- Research in HIV and Women's Health and Y This Harmony Matters (RHYTHM)
Basic Science Research Opportunities
Basic scientific research within the Division of Infectious Diseases focuses on:
- Mechanisms and control of antibiotic resistance
- Treatment of hospital-acquired infections, such as Clostridium difficile ( C. difficile )
- Pathogenesis of infectious diseases, such as bacterial meningitis
- Basic and translational research in HIV
- Immunology of host defenses and vaccine development
Major areas of investigation of bacterial pathogens include:
- Analysis of virulence mechanisms of invasive bacterial pathogens
- The role of endogenous bacterial CRISPR/Cas9 systems in host immune evasion and targeted DNA editing
- Genomics of bacterial virulence and antibiotic resistance; study of the microbiome
- Studies of the molecular mechanisms of antibiotic resistance
- Research into the role of transposable elements and repetitive nucleotide sequences in microbial pathogenesis and agents associated with bioterrorism
- Metagenomics of pathogens for basic and clinical research
The Division of Infectious Diseases participates in graduate programs and research in microbiology and molecular genetics, immunology and molecular pathogenesis, as well as population biology, ecology, and evolution.
Additional opportunities are available for basic science research in:
- Toxin-mediated infections
- Intracellular pathogens
- Hemorrhagic fever viruses
- Zoonotic pathogens
- Parasites and fungi
- Population biology
- Genomics and metagenomics
Emory Vaccine Center and Emory Hope Clinic
The Emory Vaccine Center receives more than $24 million in sponsored research funding. Primary program areas in basic science and translational research currently include HIV/AIDS, tuberculosis, malaria, Hepatitis C virus (HCV), cytomegalovirus, gamma-herpes viruses, influenza and DNA and protein-conjugate vaccines.
The Hope Clinic , a unit of the Emory Vaccine Center directed by Mark Mulligan, MD, conducts clinical research, vaccine trials and critical research on vaccine policies. A number of additional opportunities for basic scientific research are available in the Graduate Divisions of Microbiology and Immunology and through the Centers for Disease Control .
Clinical and Diagnostic Microbiology
Emory University Hospital and Grady Memorial Hospital have outstanding resources in diagnostic and clinical microbiology.
Areas of focus include:
- Development and assessment of diagnostic tools based on nucleic acid detection
- Optimization of laboratory procedures for diagnosis of mycobacterial infections
- Assessment of the accuracy of automated systems for detection of antimicrobial resistance
Clinical/Translational Research Opportunities and Global Health
Major areas of focus for clinical/translational research in infectious diseases include:
- HIV/AIDS; access to and retention in care
- Sexually transmitted infections
- C. difficile infections; prevention and treatment modalities
- Tuberculosis
- Vaccine research
- Healthcare epidemiology and healthcare infections
- Transplant Infectious Diseases
- Emerging infectious diseases
- Global health
- Bioterrorism
Clinical research training opportunities are enhanced by the presence of Clinical Interaction Sites at the Emory-affiliated hospitals .
In addition, opportunities for clinical and translational research training are enhanced by the presence of the NIH-funded CTSA award ( Georgia Clinical and Translational Science Alliance , or CTSA) which was awarded to Emory University in collaboration with Morehouse School of Medicine and the Georgia Institute of Technology. A major component of the ACTSI is the Research Education, Training and Career Development program, which includes the Master of Science in Clinical Research (MSCR).
Featured Topics
Featured series.
A series of random questions answered by Harvard experts.
Explore the Gazette
Read the latest.
Parkinson’s warning in skin biopsy

How the already anxious avoided global spike in COVID anxiety

Losing fat is good, but losing muscle isn’t
Tina Lu ’21 examined symptom internet searches early in the pandemic and scored how well they later matched clinical symptoms of COVID-19.
Kris Snibbe/Harvard Staff Photographer
Tracking progression of disease through internet searches for symptoms
Alvin Powell
Harvard Staff Writer
Student’s project found queries mirrored course of illness, foretold rise in cases
You’re not feeling well so you open a search engine and type: fever, dry cough, hoping to find hints of what you may have. A handful of days later, you’re feeling worse, and you type in: trouble breathing. It turns out you’re not the only one who’s doing this, and a Harvard senior’s research project suggests that tracking the results of all those searches can tell us something about the progression of a new disease in individuals and through a population.
Tina Lu, a Leverett House computer science concentrator, analyzed search engine data from Google Trends going back to the beginning of the coronavirus pandemic to see how well symptom searches in 32 countries on six continents matched the clinical symptoms of COVID-19 and whether the number of searches served as a harbinger of rising incidence of cases.
“We hope that our findings that this is true for COVID will be helpful for future pandemics because it might take a while for research to be done on the course of illness for a new disease, but Google Trends can be analyzed from the very beginning,” Lu said.
Published in Nature Digital Medicine earlier this year, the work found that from Jan. 1, 2020, to April 20, 2020, increases in symptom-related searches for COVID-19 preceded increases in reported cases and deaths by an average of 18.53 days. Further, the work showed that there was a clear pattern of disease progression, with early symptoms of fever, dry cough, sore throat, and chills being followed by more severe shortness of breath by 5.22 days, matching the clinical course of disease reported in medical studies.
“This is not just a one-time contribution. It’s a new tool for the public health community,” said Ben Reis, assistant professor of pediatrics at Harvard Medical School, director of the Predictive Medicine Group at the Boston Children’s Hospital Computational Health Informatics Program, and senior author on the paper. “When you’re faced with a novel pathogen, the most valuable resource is information, especially in the early stages.”
One way that modern medicine comes to understand new ailments is through case reports published in medical journals. That was the case with COVID-19, Reis said, and those observations by trained physicians will remain a valuable source of information. But journal publication can be slow, Reis said, while search engine data like that available through Google Trends can be gathered the following day or even the same day, offering a fast snapshot of what patients are experiencing and how their disease is progressing. In the case of a rapidly-moving pathogen like SARS-CoV-2, the digital data has the potential to provide invaluable early insights, Reis said.
“When a novel pandemic strikes, in addition to the existing established approaches, we propose that this could be a complementary approach to start looking at the data for different symptoms and symptom groups in any area that’s affected,” Reis said. “We’re offering an additional, complementary data source to existing sources that has the benefit of very quick availability. Google Trends or any other search engine data is available in near real time, you can get data about yesterday and in some cases, even about today.”
“We hope that our findings … will be helpful for future pandemics because it might take a while for research to be done on the course of illness for a new disease, but Google Trends can be analyzed from the very beginning.” Tina Lu
The project got underway in the months before the pandemic when Lu, then a junior, reached out to Reis at his lab at Boston Children’s Hospital. Lu was interested in applying her computer science skills to medical research and knew Reis’ lab had used Google Trends data for public health studies in the past, tracking data related to polio vaccination, for example.
The project, much of which Lu conducted from her home in New Jersey, was initially aimed at seeing whether disease peaks in different countries could be detected, but the work was confounded by different precautionary steps taken in different nations. Then she began looking at symptoms and noticed different symptoms peaked at different times in different countries. Lu looked closer and saw a clear pattern that matched what is now known as the clinical course of COVID-19.
“It was very exciting and also a little hard to completely believe at first, because I was investigating this for a really long time,” Lu said.
Lu, who was working out of her family’s basement, spent several months gathering and examining the data during the pandemic’s initial lockdown, while also juggling classwork from courses that had moved abruptly online in March 2020.
More like this

Approval of at-home tests releases a powerful pandemic-fighting weapon

A year of ‘never off’
Why some die, some survive when equally ill from COVID-19
“I was spending hours each day for many days,” Lu said. “At first, it felt like I was not seeing any consistent patterns between different countries. I would have daily calls with Ben, and we would go over different approaches or different things to look into. When we found this pattern with the course of illness, it was just very exciting because finally we found something that I guess no one else had found before and that was pretty consistent across all of these different countries we were looking at.”
Lu, who graduates this spring, plans to head to Chicago after Commencement for a job as a software engineer with an investment firm headquartered there. Though the project is over, Reis said he expects it to have a real-world impact — potentially even on variants that emerge in this pandemic. The publication outlines the methodology so others can emulate it, and he’s already fielded several inquiries from researchers.
“We’re happy to work with others. The paper was written in a way so that anyone who’s working in this space could recreate it and incorporate these approaches,” Reis said. “Our hope is that these emerging systems that have been built over the last decade to incorporate digital information into the epidemiological information cycle will incorporate these methods as well.”
Share this article
You might like.
Medical office procedure identifies key biomarker that may lead to more reliable diagnosis of neurodegenerative disorders

Psychological tools learned by those in treatment proved protective in high-stress event, study finds

Researchers call for makers of new anti-obesity drugs to study results of body composition in addition to weight loss.
So what exactly makes Taylor Swift so great?
Experts weigh in on pop superstar's cultural and financial impact as her tours and albums continue to break records.
The 20-minute workout
Pressed for time? You still have plenty of options.
Maria Ressa named 2024 Commencement speaker
Nobel Prize-winning defender of press freedom will deliver principal address
- Institute for Cardiovascular Disease Research
- Location Location
- Contact Contact
Research Projects

Cardiovascular dysfunction accounts for some of the most common and devastating diseases in the country, as well in the institute's home state of South Carolina. This research promises to uncover new understandings and breakthroughs to reduce mortality and morbidity rates due to cardiovascular disease.

Partner with Us
Become a strategic partner by starting an applied research project with us, funding the research or collaborating with us on the work.
A Holistic, Interdisciplinary Approach to Understanding Cardiovascular Disease
The Institute for Cardiovascular Disease Research is a multi-primary investigator undertaking, with highly regarded experts in the field leading novel and unique research contributions within the four-year institute plan. We seek to provide a new understanding of the ways that body inflammation plays a part in high blood pressure and identify new treatments to reduce the frequency and impact of cardiovascular disease.
High Blood Pressure Associations with Chronic, Unpredictable Stress
The chronic, unpredictable stress that comes with the everyday realities of modern life leads to widespread inflammation in the body, which causes dysfunction in the brain, vascular system, kidneys and heart along with elevated blood pressure and, eventually, cardiac failure.
Targeting the cause of inflammation, or inflammasomes, leading to cardiovascular dysfunction may uncover important high blood pressure triggers that originate in the nervous system, and open doors to new treatment strategies.
Targeted inflammasomes: NLRP3 (nucleotide-binding domain, leucine-rich–containing family, pyrin domain–containing-3) and AIM2 (absent in melanoma 2)
Focus of the institute: Neurogenic hypertension associated with chronic unpredictable stress
Research vision: A new understanding of the molecular mechanisms of inflammation in hypertension and identify new therapeutic targets to reduce morbidity and mortality in these common and devastating illnesses
Areas of Focus
By taking an interdisciplinary approach to cardiovascular health, we will advance our understanding of how stress leading to high blood pressure impacts the brain, kidneys, blood vessels and heart causes disease that ends with cardiac failure.

Project 1: Vascular System and Stress
High blood pressure and the vascular damage it does—leading to cardiovascular disease—is an enormous emotional and financial burden on patients with chronically elevated blood pressure.

Project 2: Brain and Stress
Stress has a profound impact on the brain, mood and body including causing or affecting a number of cardiovascular and other diseases and disorders. Yet, an overwhelming majority of adults report experiencing daily stress.

Project 3: Kidneys and Stress
Patient populations with higher prevalence of autoimmune disease—like women and those of the female sex, veterans and underrepresented communities of all kinds—are also at higher risk for cardiovascular disease.

Project 4: Heart and Stress
Patient populations with higher prevalence of autoimmune disease—like women and those of the female sex, veterans and those suffering from trauma and Black and African American communities—are at higher risk for cardiovascular disease.
The highest impact research today involves leveraging multiple perspectives, skillsets and areas of expertise to come up with the most comprehensive possible understanding of or solution to a significant issue.

Challenge the conventional. Create the exceptional. No Limits.

- Services Paper editing services Paper proofreading Business papers Philosophy papers Write my paper Term papers for sale Term paper help Academic term papers Buy research papers College writing services Paper writing help Student papers Original term papers Research paper help Nursing papers for sale Psychology papers Economics papers Medical papers Blog

Top 100 Disease Research Topics For Paper Writing
Students have many disease research topics to consider when writing research papers and essays. A disease occurs when the body undergoes some changes. Science philosophy has pointed at pathogens and the causes of illness. Today, medicine focus on biochemical factors, nutrition, immunology levels, and environmental toxins as the causes of diseases. Research papers on disease topics can focus on specific illnesses independently or in groups. You can also write about infectious diseases like COVID-19 and HIV or non-communicable diseases. Non-communicable diseases are also known as chronic illnesses. These are diseases that you can’t get from a sick person. They include heart disease, cancer, stroke, and lung disease. These diseases account for up to 70% of global deaths. Nevertheless, whether you opt to write about advanced topics in Lyme disease or something simple like flu, research will be paramount. You can also buy research papers cheap, if you don’t have time for it. So, d on’t put your grade at risk and get research paper online help .
Why Choose Our Disease Research Topics?
Educators want you to convince them that you have taken the time to think about your topic and research it extensively. What’s more, your research should make a meaningful contribution to your study field. Therefore, select a good topic and research it extensively before you start writing. Analyze your information to determine what will make it to your research paper. Here is a list of 100 disease research paper topics worth considering for your paper or essay.
Top Disease Research Topics
Maybe you want to research and write a research paper on a topic that anybody will find interesting to read. In that case, consider ideas in this list of disease research topics.
- How NSAIDS lead to peptic ulcers
- What are pandemic diseases?
- What is the role of pandemic diseases in the mankind history?
- What are the symptoms of acute lung disease?
- Explain how Attention Deficit Hyperactivity Disorder affects children
- Discuss the AIDS pandemic in third world countries
- Describe the main causes of AIDS
- Explain how AIDS affects children
- Discuss the treatment of AIDS
- Is alcohol addiction a disease?
- Discuss the Alzheimer’s disease scope and how it affects the elderly persons’ brain
- How can you help dementia or Alzheimer’s disease patients and caregivers?
- What are the symptoms of Alzheimer’s disease?
- What is autoimmune disease?
- Explain how autoimmune thyroiditis begins
- Examine acute protective membrane inflammation in bacterial meningitis
- Compare pathology of AIDS and black death
- Discuss the effects of cancer in today’s society
- Autism and its causes
- Different types of cancer and their prevalence
These are topics disease experts will recommend researching and writing about. And because students can write about these topics without getting complex, anybody will find them interesting. If you’re searching for research topics on Alzheimer’s disease, this list also has some ideas for you to consider.
Infectious Disease Topics for Research Papers
Are you interested in infectious disease research topics? If yes, you will find this list interesting. This category comprises hot topics in infectious disease fields. Consider some of these ideas for your research paper.
- The virology, epidemiology, and prevention of COVID-19
- The diagnosis of COVID-19
- Prevention vaccines for SARS-CoV-2 infection
- Questions people ask about COVID-19
- Clinical features of COVID-19
- COVID-19 management in a hospital setting
- Infection control for COVID-19 in homes and healthcare settings
- Skin abscess and cellulitis in adults
- Clinical manifestation, diagnosis, and epidemiology of yellow fever
- Transmission and epidemiology of measles
- Role of untreated inflammation of genital tract in HIV transmission
- Racial inequities of COVID-19 and HIV in black communities
- Community-acquired pneumonia overview in adults
- The use of procalcitonin in the infections of lower respiratory tract
- Herpes simplex virus prevention and treatment
- Uncomplicated Neisseria gonorrhea treatment
- Society guidelines for COVID-19
- Why public education is crucial in fighting COVID-19
- Overview of Ebola over the last two decades
- Investigations into the use of monoclonal antibody in treating Ebola
This category also has some of the best infectious disease presentation topics. Nevertheless, learners should prepare to research extensively before writing academic papers on these topics.
Interesting Disease Topics
Maybe you want to research and write a research paper on a topic that most people find interesting. In that case, consider these disease topics for research paper.
- Discuss bulimia as a common eating disorder
- Why are so many young people suffering from anorexia?
- What causes most eating disorders
- How serious are sleep disorders
- Discuss rabies treatment- The Milwaukee protocol
- Is assisted suicide a way to treat terminal diseases?
- What are the effects of brain injuries?
- What are professional diseases?
- Is autism a norm variant or a disease?
- The history of pandemics and epidemics
- The role of antibiotics in treating diseases
- What causes insomnia?
- What are the effects of insomnia?
- How to cope with insomnia
- Can sleeping pills cure insomnia?
- Explain what causes long-term insomnia
- Using traditional medicine to fight insomnia
- How to deal with bulimia and nervosa
- How eating disorders affect self-harm behavior
- How feminism affect anorexic women phenomenon
This is a list of easy disease topics for papers. What’s more, most people will find these research paper disease topics interesting to read about. Nevertheless, students should take time to research their preferred topics to come up with brilliant papers on any of these human disease research paper topics.
Cardiovascular Disease Research Topics
Maybe you’re interested in topic ideas on heart disease. Perhaps, you want to write about an illness of the respiratory system. In that case, consider these heart disease research topics.
- An investigation of hypertrophic cardiomyopathy
- A research of the causes of coronary artery disease
- Antithrombotic therapy in surgical valve and prosthetic heart valve repair
- Intervention choice for severe cases of calcific aortic stenosis
- Prognosis and treatment of heart failure using preserved fraction of injection
- Infective endocarditis management in adults
- Risk assessment for cardiovascular disease for primary prevention
- Prognosis and treatment of acute pericarditis
- Treatment of reflex syncope in adolescents and adults
- Anticoagulant therapy for preventing thromboembolism in atrial fibrillation
- Cardiac manifestations of COVID-19 in adults
- Acute decompensated heart failure treatment
- What is hypertriglyceridemia?
- How to manage elevated low-density lipoprotein-cholesterol in cardiovascular disease
- Management and evaluation of cardiac disease
- Conduction system and arrhythmias disease and COVID-19
- Myocardial infarction in COVID-19
- Can somebody inherit a cardiac disease?
- How effective are treatments for irregular heartbeat?
- How to determine the risk for a sudden cardiac death
This list comprises some of the best special disease topics. That’s because most people reading about these topics might not have heard about them before. Nevertheless, this category also has interesting research topics for disease control that may help individuals that want to avoid or manage some illnesses.
Research Topics for Chronic Disease
You probably know somebody living with a chronic illness. Unlike controversial topics in infectious disease, people don’t talk much about chronic illnesses. And for this reason, some people don’t know about these illnesses. When writing about non-communicable illnesses, you can settle for human genetic disease topics or even research topics for sickle cell disease. Here are some of the topics about non-communicable diseases that you can write about.
- The risk of breast cancer after childbirth
- Postpartum PTSD- Effective preventative measures
- Experiences of females suffering from cardiac disease during pregnancy- A systematic review
- Husbands attendance and knowledge of wives’ postpartum care in rural areas
- Postpartum depression screening by perinatal nurses in hospitals
- Gestational diabetes mellitus screening from the rural perspective
- Maternal mortality- How to help cardiac and pregnant patients
- Sex differences in cardio metabolic disorders and major depression- Effect of immune exposures and prenatal stress
- Determinants and prevalence of anxiety and antepartum depressive symptoms in fathers and expectant mothers- Outcomes from perinatal psychiatric morbidity
- Evaluating the effect of community health workers on non-communicable diseases, tuberculosis, malnutrition, antenatal care, and family planning
- History of women with postpartum affective disorder and the risk of future pregnancies recurrence
- New self-care guide package and its effect on neonatal and maternal results in gestational diabetes
- Depressive symptoms and life events in pregnant women- Moderating the resilience role and social support
- Gestational diabetes and ethnic disparities
- Pregnancy and diabetes- Opportunities and risks
- Cardiovascular disease maternal death reduction- A pragmatic investigation
- Meta-analysis and systematic review of gestational diabetes mellitus diagnosis with a two-step or one-step associations and approaches with negative pregnancy outcomes
- Gestational diabetes mellitus treatment in women- A Cochrane systematic overview
- Research in non-communicable diseases in Africa- A strategic investment
- How to finance the national response to non-communicable diseases
Whether you opt to write about research paper topics in Huntington’s disease or non-communicable liver disease topics, you have to engage in extensive research to come up with a brilliant paper. We have more health research topics for you, so don’t hesitate to check them. Therefore, select an idea you will be comfortable researching and writing about. That way, you will avoid enduring a boring process of investing your topic and writing the paper. If you want to hire someone to help you with your assignment, just c ontact us with a “ do my research paper now ” request and we’ll get your papers done.

Leave a Reply Cancel reply
Your email address will not be published. Required fields are marked *
Save my name, email, and website in this browser for the next time I comment.
Terms & Conditions Loyalty Program Privacy Policy Money-Back Policy
Copyright © 2013-2024 MyPaperDone.com

- Make a Gift
- Education & Training
- Global Health Security
- Emerging Infectious Diseases and Epidemics
- Medical and Public Health Preparedness and Response
- Deliberate Biological Threats
- Opportunities and Risks in the Life Sciences
- Preferred reporting items for epidemic forecasting and prediction research: the EPIFORGE 2020 statement
- Rad Resilient City Initiative
- Protecting Building Occupants from Exposure to Biological Threats
- How to Lead during Bioattacks
- Publications
- Assumptions of US PPE Needs for COVID-19
- Tips to Improve Indoor Ventilation in K-12 Schools
- Tips to make the air in your home safer during COVID-19
- COVID-19 Congressional Testimonies & Briefings
- COVID-19 Fact Sheets
- COVID-19 Situation Reports
- CommuniVax Roundups
- Testimonies & Briefings
- Post-Pandemic Recovery: From What, For Whom, and How?
- National Strategy for Improving Indoor Air Quality
- A National Conversation on Indoor Air & K-12 Schools During the COVID-19 Pandemic
- Global Forum on Scientific Advances Important to the Biological and Toxin Weapons Convention
- Events Archive
- Catastrophic Contagion
- Atlantic Storm
- Dark Winter
- Newsletter: Health Security Decoded
- Call for Papers
- The Capitol Hill Steering Committee
Disease X Medical Countermeasure Program
When will the next pandemic occur.
Any Time. Infectious disease outbreaks now occur 3 times more often than 40 years ago.
WHAT IS MOST LIKELY TO CAUSE THE NEXT PANDEMIC THREAT?
We don’t know. But, of about 2 dozen viral families capable of infecting humans, 6 families (Adenoviridae, Coronaviridae, Orthomyxoviridae, Paramyxoviridae, Picornaviridae, and Poxviridae) have these traits that will likely cause the next pandemic.
- No immunity – No preexisting immunity in the world’s population
- Airborne – Spread via respiratory transmission
- Silent – Transmissible by infected people who have no symptoms
- Harmful – No existing, effective therapeutics or vaccines
ARE WE PREPARED TO MAKE VACCINES, ANTIVIRALS, AND TESTS EVEN FASTER DURING THE NEXT PANDEMIC?
No. Currently there is $0 sustained federal funding dedicated to developing medical countermeasures for unknown viral threats.
Developing the COVID-19 vaccines in 1 year was only possible because of 15 years of prior coronavirus research + a $12 billion federal investment.
HOW CAN WE MAKE MEDICAL COUNTERMEASURES WITHOUT KNOWING WHICH DISEASE (“DISEASE X”) WILL STRIKE NEXT?
By focusing medical countermeasure development efforts on the viral families most likely to cause pandemics, rather than on a specific virus that may or may not present a future threat.
The United States should fund a new dedicated Disease X Medical Countermeasure Program that leverages technologies and vaccine platforms most suitable to the viral families that are likely to cause future catastrophic disease outbreaks. Medical countermeasures against 1 member of a viral family could easily be adapted to another member quickly when the next threat emerges. With this flexible approach, private–public partnerships could develop vaccines , antivirals , and tests for a range of unknown potential pandemic pathogens in months, not years. Stopping the next COVID-19-type pandemic a month earlier in the United States would save approximately $500 billion.
Infographic
View the full infographic below, or download the PDF here .

Additional Information
The Characteristics of Pandemic Pathogens
Expediting Development of Medical Countermeasures for Unknown Viral Threats
- Introduction to Genomics
- Educational Resources
- Policy Issues in Genomics
- The Human Genome Project
- Funding Opportunities
- Funded Programs & Projects
- Division and Program Directors
- Scientific Program Analysts
- Contact by Research Area
- News & Events
- Research Areas
- Research investigators
- Research Projects
- Clinical Research
- Data Tools & Resources
- Genomics & Medicine
- Family Health History
- For Patients & Families
- For Health Professionals
- Jobs at NHGRI
- Training at NHGRI
- Funding for Research Training
- Professional Development Programs
- NHGRI Culture
- Social Media
- Broadcast Media
- Image Gallery
- Press Resources
- Organization
- NHGRI Director
- Mission & Vision
- Policies & Guidance
- Institute Advisors
- Strategic Vision
- Leadership Initiatives
- Diversity, Equity, and Inclusion
- Partner with NHGRI
- Staff Search
Genetic Disorders
Many human diseases have a genetic component. Some of these conditions are under investigation by researchers at or associated with the National Human Genome Research Institute (NHGRI).
A genetic disorder is a disease caused in whole or in part by a change in the DNA sequence away from the normal sequence. Genetic disorders can be caused by a mutation in one gene (monogenic disorder), by mutations in multiple genes (multifactorial inheritance disorder), by a combination of gene mutations and environmental factors, or by damage to chromosomes (changes in the number or structure of entire chromosomes, the structures that carry genes). As we unlock the secrets of the human genome (the complete set of human genes), we are learning that nearly all diseases have a genetic component. Some diseases are caused by mutations that are inherited from the parents and are present in an individual at birth, like sickle cell disease. Other diseases are caused by acquired mutations in a gene or group of genes that occur during a person's life. Such mutations are not inherited from a parent, but occur either randomly or due to some environmental exposure (such as cigarette smoke). These include many cancers, as well as some forms of neurofibromatosis.
List of Genetic Disorders
This list of genetic, orphan and rare diseases is provided for informational purposes only and is by no means comprehensive.

Featured Content

Last updated: May 18, 2018
An official website of the United States government
Here's how you know
Official websites use .gov A .gov website belongs to an official government organization in the United States.
Secure .gov websites use HTTPS. A lock ( Lock Locked padlock ) or https:// means you've safely connected to the .gov website. Share sensitive information only on official, secure websites.
Biological Sciences (BIO)
- Biological Sciences (BIO) Home
- Additional Resources
- Biological Infrastructure (DBI)
- Environmental Biology (DEB)
- Emerging Frontiers (EF)
- Integrative Organismal Systems (IOS)
- Molecular and Cellular Biosciences (MCB)
- Get BIO Email Updates Get BIO Email Updates Go
- Contact BIO
- NSF on Facebook
- BIO Blog on WordPress
- Research Areas
- Biological Sciences
Multidisplinary Emerging Infectious Disease Research at NSF
BIO / CISE / EHR / ENG / GEO / MPS / OIA / OISE / SBE / TIP Interdisciplinary Partnerships / Additional Resources
This website will allow you to browse a diversity of Emerging Infectious Disease-related topic areas supported by NSF, quickly find a program that best fits your proposal idea and get contact information for Program Directors who can help you further.
* this page is best viewed using safari or firefox browser. for chrome, edge or ie (internet explorer) browsers, click here to view this page..
Emerging (and re-emerging) infectious diseases (EIDs) are occurring with increasing regularity and represent a continuing risk to the health of humans, to other animals and plants, to ecosystem health, and to economic stability. The emergence of new infectious diseases does not occur in isolation. Rather, it is closely tied to other events like interactions of pathogens with new hosts, evolutionary changes in pathogen genomes, or expansion of the geographic ranges of hosts or pathogens. Climate change, changes in land-use patterns, and human behavior all contribute to the emergence of infectious diseases.
NSF already supports crucial, multidisciplinary EID research across its Directorates; this research examines the global distribution of potential pathogens, reservoirs, vectors, and hosts; provides insight into the molecular and physical mechanisms of pathogenesis and virulence in non-human hosts; identifies host responses key to controlling or resolving infections; identifies host and microbe mechanisms that control symbiotic interactions versus pathogenic outcomes; develops a quantitative understanding of the transmission dynamics of rapidly evolving pathogens; aids in creation of sensors and methods of detecting pathogens; and investigates the cultural, social, behavioral, and economic dimensions of pathogen transmission. Finally, NSF's investment in scientific infrastructure - from supercomputers to pathogen- and ecosystem- monitoring centers - enables new knowledge and understanding of EIDs on scales not possible before.
Despite ongoing investment in EID research, scientific challenges remain including the ability to use a pathogen's genome to predict its potential transmissibility or lethality; understanding principles of host physiology and behavior, as well as the interactions between these systems and infectious agents that contribute to resistance or susceptibility to infection; understanding the underlying mechanisms responsible for an increase in pathogen host range and transmission rates, and discovering how human-built, environmental, ecological or evolutionary processes mediate these changes. If the scientific community meets these challenges, then society could more effectively predict, more quickly detect, respond to, and minimize the impact of future EIDs, thereby limiting health, economic, and other societal impacts of disease.
Sustained, cross-disciplinary research on host and pathogen biology, infection and transmission dynamics provides the basic knowledge needed to respond to and mitigate the impact of future EIDs. With a foundation in the biological sciences, success at understanding and controlling EIDs requires deep convergent integration across disciplines of science, technology, engineering, and mathematics (STEM) and effective knowledge transfer to accelerate the development of applied and translational products to mitigate EIDs including new therapeutics, sensors, monitoring and detection methods, disease models, and practices to prevent pathogen infection and spread.
To respond to these grand challenges, NSF supports EID research through core programs and special solicitations across the Directorates which include the biological, physical, and social sciences; computer and information sciences; and engineering. The core programs usually support individual research projects. Special solicitations are typically aimed at larger, more interdisciplinary, and integrative projects that go beyond the scope of what a single research program can accomplish. NSF also emphasizes support for the training of the next generation of STEM workers. By supporting young scientists, NSF enhances our nation's capacity to address EIDs and future pandemics. Descriptions of core programs and special solicitations that provide substantial support for EID research and training can be found at the links below.
Click on the Directorate / Office titles to expand.

The Directorate for Biological Sciences (BIO) is interested in funding EID research framed around principles or concepts that are generalizable. Specifically, BIO is interested in natural model systems that can be used to study EIDs. However, BIO generally does not support biological research on mechanisms of infectious disease in humans, including the etiology, diagnosis, or treatment of infectious disease or disorder. While NSF does support research into the impact of disease in animals and plants, including potential spillover of such diseases to humans, research to develop animal models of human conditions and the development or testing of procedures for their treatment is generally not supported by the Directorate of Biological Sciences.
The BIO directorate welcomes proposals in any area of research supported through the participating divisions that address the topics outlined herein. The selected topics below are of particular interest to the core programs in participating divisions. Proposals should be submitted to the core programs in the participating divisions. Questions regarding proposals should be addressed by the cognizant program officer of the program to which the proposal is to be submitted.
Click on the Division titles to expand.
Division of biological infrastructure (dbi).
- Development of software analytics or models relevant to predicting likelihood of emergence, infection and spread of infectious diseases.
- Design of novel methodologies, including bioinformatics approaches, research methods, and instrumentation, to elucidate the mechanisms of infectious disease from molecular to cellular level.
- Developing and sustaining computational platforms and tools that enable collecting, archiving, analyzing, updating, and integrating a variety of research data relevant to infectious diseases, and pathogens and their interaction with hosts.
- Creation of innovative components that integrate analytical and computational tools with a strong scientific grounding in biosciences that could lead to new discoveries or better technologies in emerging infectious disease.
- New and creative paradigm or models for training undergraduates and/or postdoctoral fellows that prepare them for a future in the emerging infection diseases.
- Postdoctoral training with sponsors in the area of emerging infectious disease, as long as they are addressing a basic science question.
- Creative use of networks of faculty and educators to address undergraduate biology education in emerging infectious diseases.
Division of Environmental Biology (DEB)
The Division of Environmental Biology (DEB) Core Track supports research and training on evolutionary and ecological processes acting at the level of populations, species, communities, and ecosystems. Research on EID related topics in the above-described areas should be discussed with a program director prior to submission to the core clusters (Ecosystem Sciences, Evolutionary Processes, Population and Community Ecology, and Systematics and Biodiversity Sciences). Additionally, EID related projects that engage quantitative, interdisciplinary, systems-oriented research on biosphere processes and their complex interactions with climate, land use, and changes in species distribution at regional to continental scales are encouraged to contact program directors in DEB's Macrosystems Biology and NEON-Enabled Science (MSB-NES): Research on Biological Systems at Regional to Continental Scales program.
Division of Integrative Organismal Systems (IOS)
- Identify elements in the host-pathogen arms race that either permit or deter establishment of infection; feed forward and feedback loops that dictate outcome or resolution of infection ( PBI , SII )
- Examine how climate change stressors (both biotic and abiotic) impacts host behavior and resilience to infection; how environmental stressors impact the development or function of the immune system to the detriment of the host ( IEP , PGRP , PBI , SII , BSC )
- Use comparative approaches to map host and pathogen gene interactions concurrently to identify genomic mechanisms responsible for disparate outcomes of infection ( EDGE , PGRP )
- Understand the role of the neural-endocrine system during infection and the effect it has on behavior changes, including those involved in sickness ( NSC )
- Understand the impact of animal behavior (e.g., sociality, species interactions, movement) as a disease spread factor and, conversely, understanding successful host-seeking behavior or behavioral manipulation by the pathogen ( BSC )
- Understand immune system development during different stages of organismal development or aging ( DSC )
- Characterize and understand the integrative context-dependent nature of infection outcomes ( All IOS ).
Division of Molecular and Cellular Biosciences (MCB)
- Examination of mechanisms that give rise to viral or other pathogen genetic variation ( GM ).
- Emergent properties of pathogens across the molecular, subcellular, and cellular scales, using laboratory, mathematical modeling, and computational approaches ( CDF , SSB ).
- Design and development of novel biological platforms that are capable of sensing and responding to emerging infectious agents ( SSB ).
- Development of new computational and experimental tools that enable the prediction of and demonstrate the limits of prediction of viral evolution ( GM , MB).
- Characterization of key biophysical properties of pathogenic virus through all atom simulations with a view to understanding function and sites of potential disruption ( MB ).
- Understanding the structure and function of potential pathogens with a view to identifying conserved regions of genomes and proteins that might be suitable as future treatment targets ( GM ).
- Understanding how host and viral genomes interact to predict potential disease outcomes as well as pathogenic range and virulence ( GM , CDF ).
- Understanding viral assembly pathways to enable disruption ( MB ).
BIO Interdisciplinary Programs
- Ecology and Evolution of Infectious Diseases (EEID)
- Predictive Intelligence for Pandemic Prevention Phase I: Development Grants (PIPP Phase I)
- Incorporating Human Behavior in Epidemiological Models (IHBEM)
Top of page

CISE Directorate Core Programs:
- Foundations of Emerging Technologies (FET) Cluster
- Information Integration and Informatics (III) Cluster
Smart Health and Biomedical Research in the Era of Artificial Intelligence and Advanced Data Science (SCH):
The purpose of this interagency program with the National Institutes of Health is to support the development of transformative high-risk, high-reward advances in computer and information science, engineering, mathematics, statistics, behavioral and/or cognitive research to address pressing questions in the biomedical and public health communities. Transformations hinge on scientific and engineering innovations by interdisciplinary teams that develop novel methods to intuitively and intelligently collect, sense, connect, analyze and interpret data from individuals, devices and systems to enable discovery and optimize health. Solutions to these complex biomedical or public health problems demand the formation of interdisciplinary teams that are ready to address these issues, while advancing fundamental science and engineering.
Addressing the challenges will require fundamental research and development of new tools, workflows and methods across many dimensions; some of the themes are highlighted below.
- Information Infrastructure: This theme encourages pursuit of fundamental research in data management to enable interoperable, distributed, federated, and scalable digital infrastructure to optimize knowledge discovery.
- Transformative Data Science: This theme supports development of novel computational approaches for fusion and analysis of multi-level and multi-scale clinical, imaging, biomedical, personal, behavioral, social, contextual, environmental, and organizational data to maximize inferences that can be derived from the data.
- Novel multimodal sensor system hardware: This theme addresses the need for new multimodal sensing systems/platforms and analytics to generate predictive and personalized models of health.
- Effective Usability: To generate technology that is usable and effective will require development of new approaches, taking into account ethical, behavioral and social considerations, to support individuals to effectively participate in their own health, diagnosis, and treatment, such as personalized information systems, personalized and dynamic treatment, accessing and visualizing health data and knowledge that support users across socio-economic status, digital and health literacy, technology and broadband access, geography, gender, and ethnicity.
- Automating Health: This theme supports work that enables interoperable, temporally synchronized, devices and systems to connect data and devices and create closed-loop or human-in-the-loop systems to assess, treat and reduce adverse health events.
- Medical image interpretation. This theme's goal is to determine how characteristics of human pattern recognition, visual search, perceptual learning, attentional biases, etc. can inform and improve use and development of presentation modalities (e.g., pathologists reading optical slides through a microscope vs. digital whole-slide imagery) and identify the sources of inter- and intra-observer variability in medical image interpretation.
- Unpacking health disparities. In this theme, proposals should seek to develop holistic, data-driven or mathematical models to address the structural and/or social determinants of health. The above listed themes are to provide examples for possible research activities that may be supported by this solicitation, but by no means should the proposed research activities be restricted to these themes. These research themes are clearly not mutually exclusive, and a given project may address multiple themes.
CISE Interdisciplinary Programs
- Collaborative Research in Computational Research in Neuroscience (CRCNS)
- Integrative Strategies for Understanding Neural and Cognitive Systems (NCS)
- Semiconductor Synthetic Biology Program (SemiSynBio)
- Smart Health and Biomedical Research in the Era of Artificial Intelligence and Advanced Data Science (SCH)

The Directorate for Education and Human Resources (EHR) supports research and workforce development programs that contribute to the growth of the U.S. bioeconomy. EHR invests in foundational and future-oriented STEM educational research as well as innovative educational programs that enable students to enter and pursue careers in advanced technologies. EHR places priority on investments that support broadening participation and build a diverse, highly skilled U.S. STEM workforce and a STEM-literate public.

Click on the title to expand.
Eng interdisciplinary programs.
- Smart Health and Biomedical Research in the Era of Artificial Intelligence and Advanced Data Science (SCH )

GEO Interdisciplinary Programs

Division of Chemistry (CHE)
The Division of Chemistry (CHE) supports fundamental chemistry research that focuses on development of new chemical sensing and imaging tools for diagnostics or monitoring of bacterial and viral infections, as well as of novel approaches for understanding the underpinning molecular mechanisms and subsequent mitigating strategies against EIDs and future pandemics. For example, the successful development of mRNA vaccines has required and will continue to require fundamental research in unnatural nucleic acids as well the use of custom lipid synthesis for storage and biological delivery. For this meta-program, the most relevant programs in the CHE Division are: Chemical Measurement and Imaging (CMI), Chemistry of Life Processes (CLP), and Chemical Synthesis (SYN).
Participating Program description:
Chemical Measurement and Imaging (CMI) : The Chemical Measurement and Imaging Program supports work advancing new measurement science enabling capabilities likely to be of use to the chemistry community. New approaches and/or improved understanding of important existing approaches are supported. Envisioned applications - including health-related applications - can contribute to the broader impacts but should not be the primary focus. Adaptation and/or optimization of existing methods for specific applications should be directed elsewhere.
Chemistry of Life Processes (CLP) : The Chemistry of Life Processes Program supports fundamental research at the interface of chemistry and biology that advances the understanding of the molecular underpinnings of life processes. These processes include those involved in the emergence and transmission of infectious diseases and in disease detection, diagnostics, and therapeutics. For example, new chemical probes and characterization strategies can be applied to the study of the structure and dynamics of viral RNA and of proteins and enzymes required for viral infectivity and replication, which in turn can provide target sites for new therapeutics.
Chemical Synthesis (SYN) : The Chemical Synthesis (SYN) program in the Division of Chemistry encourages submission of proposals that target the synthesis of novel bioactive molecules, including metalloantibiotics, with potential for applications in infectious disease research. Such projects may involve the development of new methodology for the assembly of chemical structures for selective biorecognition, utilization of chemoenzymatic approaches for elaboration of bioactive compounds, and design of organic compounds and/ormetalloantibiotics that target specific cell membranes. Collaborative proposals with multiple PIs that partner fundamental chemical synthesis with programs in infectious disease research are especially encouraged.
Divisipon of Mathematical Sciences (DMS)
The Mathematical Biology program in the Division of Mathematical Sciences (DMS) supports many research projects closely related to EID including mathematical modeling the development of innovative mathematical frameworks of emerging and reemerging infectious diseases. Some of these projects involve close collaborations of mathematicians with epidemiologists, immunologists, public health professionals and policymakers. Specific research topics range from short-term forecast of disease outbreaks to fundamental understanding of long-term behavior of pathogen evolution and disease characteristics. Other topics include cell-pathogen interactions at within-host level to host-pathogen dynamics and transmission at population and community levels. Additionally, some projects involve coupling of biological processes at multiple temporal and spatial scales, using mathematical/statistical/computational tools and theories with various modeling approaches most appropriate for addressing the biological questions. Also, recent projects include potentially transformative research concerning the incorporation of human social, behavioral, and economic processes into mathematical epidemiological models.
Other programs within DMS, including Statistics and Computational Mathematics, also support research projects pertaining to EID.
DMS is involved in the EEID (Ecology and Evolution of Infectious Diseases) program and plans to co-fund research projects that include strong mathematical components for addressing important questions related to modeling of infectious diseases. The joint DMS/NIGMS initiative also supports research related to infectious diseases that may lead to a more general understanding of the underlying problems and have broader applicability (as opposed to applications to a specific disease).
The programs mentioned above focus on individual or small group research projects. A new solicitation in which DMS plays a key role, National Institute for Theory and Mathematics in Biology (NITMB), aims at larger and more interdisciplinary research activities. One of the topics for this solicitation is Infectious Pathogens: Immunology and Transmission. The research topics include modeling the evolutionary outcomes of host-pathogen interactions as dynamically coupled immunological and epidemiological processes. This is a grand challenge, particularly when models incorporate heterogeneities in attributes such as pathogen virulence, host defense mechanisms, and human intervention. New mathematical theory and modeling approaches are needed for exploring emergent properties of infectious pathogens, uncovering principles in immunology, and predicting effects on pathogen transmission dynamics, especially in the presence of multiple scales of time, space, and biological processes within changing environments.
MPS Interdisciplinary Programs

Click on the Office titles to expand.
Integrative activities.
- Integrative Activities administers NSF-wide programs such as Science and Technology Centers, Major Research Instrumentation, Mid-scale Research Infrastructure - Track 1, and Historically Black Colleges and Universities - Excellence in Research
Established Program to Stimulate Competitive Research (EPSCoR)
- The Established Program to Stimulate Competitive Research (EPSCoR) Section enhances research competitiveness of targeted jurisdictions (states, territories, commonwealth) by strengthening STEM capacity
Evaluation and Assessment Capability (EAC)
- The Evaluation and Assessment Capability (EAC) Section, which provides centralized support and resources for data collection, analytics, and the design of evaluation studies and surveys. These activities enable NSF to more consistently evaluate the impacts of its investments, to make more data-driven decisions, and to establish a culture of evidence-based planning and policymaking.

The Office of International Science and Engineering (OISE) supports international research and research-related activities for U.S. science and engineering students through its International Research Experiences for Students (IRES) program and strategic linkages between U.S. and international groups through its Accelerating Research through International Network-to-Network Collaborations (AccelNet) program. These networks leverage research and educational resources to tackle grand research challenges that require significant coordinated international efforts and contribute the needed collaborations and are essential to advancing and expanding the bioeconomy.
- NSF plans to launch Global Centers , an international center-level activity that will enable interdisciplinary and international teams to address grand societal challenges through use-inspired research. NSF anticipates releasing a funding opportunity in FY 2022 on topics related to climate change and clean energy and is developing Global Center-related partnerships with international counterpart funding agencies.
- Partnerships in International Research Experiences (PIRE) funds international research collaborations between U.S.-based investigators and researchers in foreign countries. The current solicitation is for Use Inspired Climate Change and Clean Energy Research Challenges.

The Directorate for Social, Behavioral and Economic Sciences (SBE) sciences focuses on human behavior and social organizations, including how social, economic, political, cultural, and environmental forces affect the lives of people from birth to old age and how people in turn shape those forces. SBE scientists develop and employ rigorous methods to discover fundamental principles of human behavior at levels ranging from cells to society, from neurons to neighborhoods, and across space and time.The SBE research divisions (Behavioral and Cognitive Sciences, Social and Economic Sciences) include programs that focus on cultural and social systems, human biology and geography, psychology and cognition, language, economic and political systems, and even the scientific enterprise itself.
Fundamental research on human behavior can provide unique and critical insights about the ways humans influence, and are influenced by, host and pathogen biology and infection and transmission dynamics, thereby informing and improving our ability to effectively investigate, monitor and respond to future emerging infectious diseases (EIDs). In addition, the SBE sciences can provide insight into concurrent and long-term social issues associated with EIDs. Though individual SBE programs do not focus exclusively on EID research topics, individual project ideas addressing aspects of human behavior in the context of EIDs may be relevant to individual SBE programs. Questions regarding proposals should be addressed by the cognizant program officer of the SBE program to which the proposal is to be submitted.
SBE Interdisciplinary Programs

The Directorate for Technology, Innovation and Partnerships (TIP) harnesses the nation's vast and diverse talent pool to advance critical and emerging technologies, address pressing societal and economic challenges and accelerate the translation of research results from lab to market and society. TIP improves U.S. competitiveness by growing the U.S. economy and training a diverse workforce for future, high-wage jobs.
TIP programs, Convergence Accelerator and Regional Innovation Engines and Partnerships Enabling Open Source Ecosystems foster innovation and technology ecosystems. Programs that are part of the Lab to Market platform, Partnerships for Innovation , I-Corps , Small Business Innovation Research (SBIR) and Small Business Technology Transfer (STTR) , establish translation pathways. All programs support biotechnology applications that serve to build a vibrant and inclusive U.S. bioeconomy.

Interdisciplinary Programs
- Dynamics of Integrated Socio-Environmental Systems (DISES)
- Plant-Biotic Interactions
- Symbiosis, Infection, and Immunity
- Predictive Intelligence for Pandemic Prevention (PIPP)

- Português Br
- Journalist Pass
Medical Research
- AI and Digital Health
- Biotherapeutics
- Clinical Trials
- Discovery Science
- Healthcare Delivery
- Individualized Medicine
- Translational Science

Mayo Clinic research finds a connection between Social Network Index score and AI-determined biological age ROCHESTER, Minn. — A new study from Mayo Clinic finds[...]
Latest stories

Signup to receive email notifications
Explore more topics

For Journalists Only Sign up for a Journalist Pass

Disease: Research Project Outline
- Categories: Alzheimer's Disease Health Care Reform
About this sample

Words: 409 |
Published: Mar 20, 2024
Words: 409 | Page: 1 | 3 min read
Introduction
- Understanding the pathological processes involved in Alzheimer's disease
- Identifying potential biomarkers for early diagnosis and disease progression
- Exploring novel therapeutic targets for the treatment of Alzheimer's disease

Cite this Essay
Let us write you an essay from scratch
- 450+ experts on 30 subjects ready to help
- Custom essay delivered in as few as 3 hours
Get high-quality help

Verified writer
- Expert in: Nursing & Health Government & Politics

+ 120 experts online
By clicking “Check Writers’ Offers”, you agree to our terms of service and privacy policy . We’ll occasionally send you promo and account related email
No need to pay just yet!
Related Essays
3 pages / 1423 words
4 pages / 1991 words
3 pages / 1438 words
1 pages / 1282 words
Remember! This is just a sample.
You can get your custom paper by one of our expert writers.
121 writers online
Still can’t find what you need?
Browse our vast selection of original essay samples, each expertly formatted and styled
Related Essays on Alzheimer's Disease
Alzheimer's disease is a devastating neurodegenerative disorder that has become increasingly common in recent years. The disease primarily affects elderly individuals, and it is estimated that nearly six million people in the [...]
Alzheimer's disease is a debilitating condition that affects millions of people worldwide. It is a progressive disorder that gradually destroys memory, thinking skills, and eventually the ability to carry out even the simplest [...]
Alzheimer's disease, a neurodegenerative brain disease, is the most common cause of dementia. It currently afflicts about 4 million Americans and is the fourth leading cause of death in the United States. Furthermore, [...]
Neurodegenerative diseases are becoming more prominent in today’s world. Be it due to life style changes or due to inheritance… the number of cases in this domain of medicine are on constant rise. The most prevalent of them are [...]
The Curious Incident of the Dog in the Night-Time introduces fifteen-year-old Christopher Boone, whose counselor has suggested that he write a book. Christopher's book is about his quest to find out who murdered his neighbors' [...]
Autism Spectrum Disorder (ASD) encompasses a wide range of social and mental afflictions that are difficult to treat. Due to a lack of established treatments for ASD, alternative therapies have been the primary form of [...]
Related Topics
By clicking “Send”, you agree to our Terms of service and Privacy statement . We will occasionally send you account related emails.
Where do you want us to send this sample?
By clicking “Continue”, you agree to our terms of service and privacy policy.
Be careful. This essay is not unique
This essay was donated by a student and is likely to have been used and submitted before
Download this Sample
Free samples may contain mistakes and not unique parts
Sorry, we could not paraphrase this essay. Our professional writers can rewrite it and get you a unique paper.
Please check your inbox.
We can write you a custom essay that will follow your exact instructions and meet the deadlines. Let's fix your grades together!
Get Your Personalized Essay in 3 Hours or Less!
We use cookies to personalyze your web-site experience. By continuing we’ll assume you board with our cookie policy .
- Instructions Followed To The Letter
- Deadlines Met At Every Stage
- Unique And Plagiarism Free

Scientists take on 'moonshot' project mapping human brain to help fight disease
To many other researchers, creating a map of the 86 billion neurons, or nerve cells, that make up the brain would be considered a nearly insurmountable project. However, researchers at the Allen Institute are pursuing that very task, saying it will lay the foundation for helping to better understand how the brain functions the way it does.
The Allen Institute, founded by Microsoft co-founder and philanthropist Paul Allen in 2003, was originally created to map gene activity in the mouse brain, but researchers quickly began including studies of the human brain in their work. The non-profit is the workplace of hundreds of researchers who, in large teams, launch aspirational ‘moonshot’ projects that could help solve the biggest questions regarding diseases afflicting millions of Americans.
"It does things a bit differently than other research institutions," Dr. Rui Costa, president and CEO of the Allen Institute, told ABC News. "We do big moonshot projects in life sciences that last sometimes 10, 15 years, where teams of interdisciplinary scientists answer a specific question, and then we share openly with the world."
Costa says these projects lay foundational work that can be used by other institutions. However, they are extremely complex and need to be done to scale, which requires either a multidisciplinary team, or even larger consortiums.
Dr. Ed Lein, senior investigator at the Allen Institute for Brain Science, said it was only recently that the tools existed to enable researchers to make high-resolution maps of the brain, which in turn can provide complete descriptions of all the cells that make up the organ.
He says the maps, described as a moonshot project, will be akin to sequencing a genome, which – when complete – describes a person’s complete genetic makeup.
"That becomes a really powerful reference for not only understanding the normal brain but understanding disease," Lein told ABC News. "So, we also have major efforts in trying to understand Alzheimer's disease, and the really specific types of cells that may be vulnerable in disease, as well as trying to create new tools that can target particular cell types that may actually be useful for gene therapies. "
"For example, if we can identify the vulnerable types of cells, we can now develop tools to check to start to target them,” Lein noted. “And so, it's a whole new research area that's been opened up by new technologies."
There are hundreds of brain diseases, meaning millions of American suffer from them. Alzheimer's disease, which is the most common type of dementia, affects about 6.7 million people . Parkinson's disease, a progressive disorder caused by degeneration of nerve cells in the part of the brain that controls movement, is expected to affect 1.2 million people by 2030 .
Neither Alzheimer's nor Parkinson's disease has a cure, and most treatments focus on managing symptoms and maintaining quality of life. The Allen Institute team hopes its research can change that, and potentially lead to targeted therapies that help eliminate the diseases.
"To develop any type of pharmacologies or pharmacotherapeutics, or even understand the way diseases work and how they are damaging to the brain and cell types, we have to understand what are the basic core components of the cell type," Dr. Brian Lee, a senior scientist in the integrative cell physiology department at the Allen Institute, told ABC News. "So, this provides that information for people to start to do some targeted approaches to fixing or treating Alzheimer's or Parkinson's."
Lee explained that in the case of Parkinson's disease, there is a loss of cells in the basal ganglia circuitry , located near the center of the brain and which are responsible for controlling movement in the body.
However, it's unclear which types of cells are being affected by disease. A map of the brain’s cells could help answer this question.
"So, it's going to be super helpful, the more that we start to understand all the cell types in the brain and how different cell types are affected to disease, we can start taking a more a very more selected and targeted approach to treating it and, hopefully, eliminating it," Lee said.
Lein said similarly, for Alzheimer's, research has shown that specific types of cells are affected at various points in the disease’s progression and in different regions of the brain. He said the brain map can be used to map data on Alzheimer's to determine if and how these neuronal losses are leading to cognitive decline.
"I think this is just the beginning of this field so just like the genome, these maps will serve this role that now anyone studying any disease can come and get this high-resolution perspective on what's happening in that disease," Lein said. "And so, it's sort of a big community moonshot here, but it begins with this fundamental foundational atlas."
The Allen Institute is also working on other ways to try and get a better understanding of the brain. For example, Lee is working on a project that allows researchers to essentially probe or poke a single neuron to understand its functional properties. The cell is also filled with a dye to further determine its shape and function. Because there are thousands of cell types in the brain, all doing vastly different, complex tasks, Lee said this helps understand how each cell contributes to the brain’s make-up and function.
He added that the institute has also collaborated with local neurosurgeons who often must remove healthy brain tissue from patients during surgery, such as in cancer patients to reach a brain tumor. Researchers at the institute then collect some of those tissues and use the finite time they have while the tissues are alive to culture them and study individual neurons.
Allen Institute CEO Dr. Rui Costa said this research is not only setting a framework for a further and deeper understanding of the brain, but it’s also providing answers to what makes humans unique creatures.
"To understand the brain is something very important for humans, because the brain is essential to who we are, what we love, what we hate, what we remember, how we behave, how we feel," he said. "Everything that makes our persona, in a way, depends on how our brains evolve, and they experience things throughout life. So, to understand the brain is one of the big quests of humanity."

MTA Missing Math Clock
Release: as part of new “project women’s health” initiative, gottheimer announces legislation to combat rare diseases impacting women.
Multi-front effort to boost clinical trials and find treatments and cures
Women often left out or underrepresented in clinical trials

Above: Gottheimer at Cooperman Barnabas Medical Center.
LIVINGSTON, NJ — Today, March 19, 2024, U.S. Congressman Josh Gottheimer (NJ-5) announced new legislation, the Securing Equal Access to Research, Care, and Health, or SEARCH Act, to help fund new efforts to increase the number of women in clinical research trials, leading to treatments and cures for rare diseases and blood disorders which affect millions of women in New Jersey and across our nation. The legislation is part of Gottheimer’s new, multi-front legislative initiative: “Project Women’s Health.”
Video of the announcement can be found here .
Gottheimer also highlighted his support for the President’s State of the Union Address earlier this month, which called on Congress to provide $12 billion to research health conditions and diseases that disproportionately affect women. In Congress, Gottheimer plans to work to help make this investment a reality.
“There is a clear gap in research, awareness, diagnosis, management, treatment, and cures of diseases and disorders affecting women. We must address the inequality between men and women in research, clinical trials, and awareness. It must be taken seriously and fixed and there’s no better time than the present,” said Congressman Josh Gottheimer (NJ-5), Rare Disease Caucus Member. “Even with all of the challenges we are facing, I remain optimistic and hopeful. I hope that you are, too. Built on collaborative work with experts and advocates, I’m confident that the SEARCH Act will ultimately lead to life changing medication, treatment, and cures for women impacted by rare diseases and bleeding disorders.”
“The Congressman is an advocate for ensuring equal access to care for all residents in New Jersey. He is focused on addressing disparities and improving healthcare outcomes across the state. We share a deep commitment with the Congressman to the most vulnerable among us,” said RWJBarnabas Health President and CEO Mark E. Manigan. “While we’re very proud of the awesome clinical services we offer here at Cooper and across the system, we are equally proud of that commitment to the most vulnerable as this state’s largest provider of charity care and as the state’s largest providers of care to beneficiaries to the medicare program by two times.”
“Thank you congressman for giving me the opportunity to speak this morning and for leading the charge on the SEARCH Act. It’s an important piece of legislation that champions the health of women, especially those affected by rare disease. The importance of the SEARCH act goes beyond my professional role and in fact connects deeply and is personal to me. In 1999, my family’s life changed forever when my niece who was ten years old at the time was diagnosed with Friedreich’s ataxia — a rare, debilitating, degenerative, neuromuscular disease. Sadly, in 2020, my niece’s battle ended just months after her 30th birthday… just last year… almost three years to the day after she died, the FDA approved a new medication to slow the progression for Friedreich’s ataxia. A breakthrough that arrived too late for my niece, but marked a significant step forward for others. The SEARCH Act represents a beacon of hope for women suffering from rare diseases,” said RWJBarnabas Health Vice President of Women’s Services Suzanne Spernal.
“I have witnessed the disparities women face and have faced for decades. Women are overlooked, misdiagnosed, or not diagnosed at all and it has become clear that legislation of this kind is not only necessary but critical for women, not only with bleeding disorders but also for women in the rare disease community at large,” said Hemophilia Association of New Jersey Executive Director Stephanie Lapidow.
“Although a disease may be rare, thus impacting a small number of people, it does not change the profound impact a condition has on those afflicted, as well as their families. Not uncommonly, due to the limited understanding of these diseases and the small number of people affected, funding for research is often scarce, making it difficult to find treatments,” said RWJBarnabas Director of Cell Therapy and Bone Marrow Transplantation Northern Regions Dr. Adrienne Phillips . “For the individual or family with a disease, getting to a diagnosis may be a long and frustrating road fraught with prolonged diagnosis and treatment delays.”
Challenges Facing Women’s Health Related to Rare Diseases and Blood Disorders:
- Since 1994, fewer than 4% of adults in the United States participate in clinical trials despite increasingly prolonged recruitment periods.
- Up to 85% of clinical trials fail to recruit or retain a sufficient sample size, leading to failures to meet targets in four out of every five trials. For rare diseases more commonly impacting women, like rett syndrome and multiple sclerosis, it can be even harder to start and maintain proper trials.
- The CDC estimates that up to 1 in 100 women and girls in the United States — more than 1.5 million Americans — have a bleeding disorder, many of whom aren’t even aware of their condition.
- The most common bleeding disorder affecting women is von Willebrand disease, or VWD, which results from a deficiency or defect in the body’s ability to produce a certain protein that helps blood clot.
- Reports indicate that women are directed to a hospital and specialists later than men following the onset of symptoms, which delays diagnosis and care. This can often lead to a rapid progression of the disease.
Gottheimer’s New Legislation, the Securing Equal Access to Research, Care, and Health or SEARCH Act will:
- There are many diseases that disproportionately impact women and are often swept under the research and funding mat.
- Fund recruitment campaigns for women in NIH clinical research trials for rare diseases by increasing advertising at hospitals, doctors’ offices, health centers, and healthcare clinics. This can have a direct and immediate impact on helping Jersey families.
- Require the CDC and HHS to increase the number of women in federal bleeding disorder programs that provide funding for research, surveillance, prevention, and services through public awareness campaigns.
- Create a task force between the HHS, NIH, FDA, CMS, and the private sector , including hospitals and labs in New Jersey, to produce a report on the rare diseases that disproportionately impact women, helping ensure more funding goes toward diseases and conditions that impact women.
Gottheimer was joined by RWJBarnabas Health President and CEO Mark E. Manigan, Cooperman Barnabas Medical Center President and CEO Richard L. Davis, RWJBarnabas Health Senior Vice President Dr. Balpreet Grewal-Virk, RWJBarnabas Health Vice President of Women’s Services, Director of Cell Therapy and Bone Marrow Transplantation Northern Regions Adrienne Phillips, and Hemophilia Association of New Jersey Executive Director Stephanie Lapidow.
Below: Gottheimer with rare disease doctors, nurses, and advocates.

Gottheimer’s remarks as prepared for delivery:
Good afternoon, and thank you again to Mark and Rick, Dr. Phillips, Stephanie, and Suzanne. It’s great to be back here in Livingston at Cooperman Barnabas Medical Center. I say back because I was born here and grew up down the road in North Caldwell. I also had surgery here after I broke my elbow as a kid. I can’t tell you the exact room. It’s a little different than what I remember — and the hospital looks fantastic. I do remember my mom sneaking me apple pancakes from the Ritz diner.
Before I begin, I want to thank all our nurses, doctors, frontline health care workers, our great North Jersey bio and life sciences leaders, and of course our rare disease researchers and advocates for the incredible work they do each and every day for our families. I cannot thank all of you enough for everything you do for our community to help improve the health and lives of countless Americans.
There are a few reasons it’s appropriate that we’re here this morning. March not only is it Women’s History Month, but it’s Blood Disorder Month, and it was just recently Rare Disease week.
Just yesterday, recognizing the importance of this month, the President signed an Executive order directing federal agencies to deploy federal funds to research health conditions and diseases that disproportionately affect women. As a member of the Rare Disease Caucus in Congress, I couldn’t agree more.
That is why today, I’m here to announce new legislation to help fund a new effort to increase the number of women in clinical research trials, leading to treatments and cures for rare diseases and blood disorders which affect millions of women across our nation.
It’s all part of my new, multi-front legislative initiative project women’s health which builds on my years of work on this front.
When it comes to research on rare and blood diseases, including those that disproportionately affect women, there is a shortage of women in clinical trials. My new legislation — the Securing Equal Access to Research, Care, and Health or SEARCH Act — will take that on.
As a quick refresher, a rare disease is one that impacts fewer than 200,000 people and far too often has no FDA-approved treatments. We just heard from Suzanne about her niece who was diagnosed with Friedreich Ataxia and sadly passed away at the age of 30, just three years before an FDA medication was approved to help delay the effects of her condition. As Suzanne mentioned, if we had had more investments for recruitment and public awareness campaigns for clinical trials, maybe the medication to delay the effects of her niece’s condition would have been discovered in time.
Personally, I lost my mom, of blessed memory, to a rare disease, Sarcoidosis, four and a half years ago.
While we are making progress, and have had some big breakthroughs, it’s far too slow, and we still have a long way to go. In fact, 95 percent of the more than 10,000 rare diseases that impact more than 30 million Americans have no FDA-approved treatment.
Rare disease research is critical. Not only can it lead to a cure or treatment for those afflicted, but the research can also be a gateway to breakthroughs for other treatments and cures.
Here in New Jersey, we are lucky to have some of the best, hospitals and medical facilities, labs, and R&D in the country, the best doctors, medical school graduates, nurses, and life sciences leaders.
With fourteen of the world’s top twenty research-based biopharmaceutical companies here in our state, we not only have cutting-edge hospitals, but our pharmaceutical and medical device companies have led the way on some of the biggest medical breakthroughs. New Jersey ranks second in the nation for cancer medicines in development and first for heart and stroke drugs and development. In 2017, 50 percent of all new FDA approvals came from companies with a New Jersey footprint.
What’s great is we already have some success stories from rare disease and bleeding disorder research.
In 2019, there were new approvals for the first triple combination therapy for patients with cystic fibrosis and a new gene therapy to treat pediatric patients under two-years-old with spinal muscular atrophy.
And just last year, the FDA approved gene therapies for hemophilia A and hemophilia B — an important advancement in providing treatment options and reducing the need for ongoing, routine therapy.
And Allergan, right here in New Jersey, announced FDA approval for an existing drug application that can now be used to treat lower-limb spasticity in children ages two to seventeen.
I want us to double down, so that we expand in the years ahead. We can lead the way in helping to develop the next cures for orphan diseases and blood disorders too commonly leaving women without the support and treatments they need.
And when we find those cures, it’ll help the amazing medical professionals — like here at Cooperman Barnabas Medical Center — care for women too often left without the help they need.
As Suzanne intimated, there are clear challenges and obstacles — especially when it comes to research.
First, participation in clinical trials is poor. Since 1994, fewer than four percent of adults in the United States participate in clinical trials despite increasingly prolonged recruitment periods. What’s worse, up to 85 percent of clinical trials fail to recruit or retain a sufficient sample size, leading to failures to meet targets in four out of every five trials. For rare diseases more commonly impacting women, like rett syndrome and multiple sclerosis, it can be even harder to start and maintain proper trials.
Second, like rare diseases, women with bleeding disorders have often been underrecognized and underdiagnosed. The CDC even estimates that up to 1 of every 100 women and girls in the United States — more than 1.5 million Americans — have a bleeding disorder, many of whom aren’t even aware of their condition. The most common bleeding disorder affecting women is von Willebrand disease, or VWD, which results from a deficiency or defect in the body’s ability to produce a certain protein that helps blood clot. Women with bleeding disorders have historically been excluded from research.
Third, because women tend to have rare diseases diagnosed later than men, bias in cures or treatments start early and leave lasting impacts. Reports indicate that women are directed to a hospital and specialists later than men following the onset of symptoms, which delays diagnosis and care. This can often lead to a rapid progression of the disease.
The bottom line: There is a clear gap in research, awareness, diagnosis, management, treatment, and cures of diseases and disorders affecting women. It must be taken seriously and fixed. We must do more to provide hope and support to the families who struggle every day to manage a rare disease or bleeding disorder — or any disease, for that matter. We must address the inequality between men and women in research, clinical trials, and awareness. And there’s no better time than the present.
That’s why, today, as I mentioned earlier, I’m here with medical professionals, advocates, and family members of those taken from rare diseases to announce the Securing Equal Access to Research, Care, and Health or SEARCH Act. This new legislation I’m introducing in Congress will support new research and clinical studies — and hopefully new treatments and cures — for diseases impacting women, including rare diseases and blood disorders.
First, the legislation will require the National Institute of Health, or NIH, to produce an action plan within 180 days to highlight the rare disease and health complications that uniquely impact women. As we laid out today, there are many diseases like blood disorders that disproportionately impact women and are often swept under the research and funding mat.
Second, the SEARCH Act will fund recruitment campaigns for women in NIH clinical research trials for rare diseases by increasing advertising at hospitals, doctors’ offices, health centers, and healthcare clinics. This can have a direct and immediate impact on helping Jersey families.
Third, the new legislation requires the CDC and HHS to increase the number of women in federal bleeding disorder programs that provide funding for research, surveillance, prevention, and services through public awareness campaigns.
Fourth, the SEARCH Act will create a task force between the HHS, NIH, FDA, CMS, and the private sector, including hospital and labs in New Jersey, to produce a report on the rare diseases that disproportionately impact women, helping ensure more funding goes toward diseases and conditions that impact women.
Finally, in his State of the Union Address earlier this month, the President called on Congress to provide $12 billion to research health conditions and diseases that disproportionately affect women. In Congress, I plan to work overtime in the coming months to make this investment a reality. We must pass legislation to create a new fund for Women’s Health Research at the NIH that will support a nationwide network of cutting-edge research centers in women’s health care.
Today, we’re here as part of my new, legislative fight on multiple fronts to protect women’s health care – whether that’s about research, clinical trials, treatments, and finding cures, or doing everything we can to protect a woman’s fundamental right to make her own personal, private healthcare decisions.
Even with all of the challenges we are facing, I remain optimistic and hopeful. I hope that you are, too. Built on collaborative work with experts and advocates, I’m confident that the SEARCH Act will ultimately lead to life changing medication, treatment, and cures for women impacted by rare diseases and bleeding disorders.
In the greatest country in the world, if we can come together to solve problems and support women’s health, I know that our best days will always be ahead of us. Once again, thank you for joining me here today.
May God bless you, your families, and may God continue to bless the United States of America.
Recent Posts
Release: gottheimer, kean lead bipartisan letter thanking germany for supporting israel’s right to self-defense.
WASHINGTON, D.C. — U.S. Representatives Josh Gottheimer (NJ-5) and Tom Kean (NJ-7) led a letter signed by a bipartisan group of more than twenty Members of Congress thanking German Chancellor Olaf Scholz for announcing his country’s intention to intervene on Israel’s behalf and for supporting Israel’s right to self-defense in the International Court of Justice […]
RELEASE: Gottheimer-led Bipartisan Legislation Overwhelmingly Passes House to Protect Americans from TikTok
Controlled by Chinese Government Disinformation and Data Mining Machine Used to Undermine Our Democracy WASHINGTON, D.C. — Today, March 13, 2024, bipartisan legislation co-led by U.S. Representative Josh Gottheimer (NJ-5), with Representative Mike Gallagher (WI-8) and Raja Krishnamoorthi (IL-8), and the House Select Committee on the Strategic Competition Between the United States and the Chinese Communist […]
RELEASE: In Legislation Signed Saturday, Gottheimer Secures New Federal Dollars for North Jersey Communities and Families
Claws Back More Federal Dollars from Washington to the Fifth District to Help Invest and Protect Towns, Lower Taxes WASHINGTON, D.C. — Today, March 12, 2024, thanks to new legislation passed out of the House and signed into law on Saturday, U.S. Congressman Josh Gottheimer (NJ-5) announced more than $10 million in new federal investments […]
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Orphanet J Rare Dis

Scientific evidence based rare disease research discovery with research funding data in knowledge graph
1 Division of Pre-Clinical Innovation, National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (NIH), Rockville, MD 20850 USA
Ðắc-Trung Nguyễn
Timothy sheils, gioconda alyea.
2 ICF International Inc, Rockville, MD USA
3 Office of Rare Diseases Research, National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (NIH), Bethesda, MD 20892 USA
James Dickens
Ewy a. mathé, anne pariser, associated data.
The data applied in this study can be accessed at http://grants4rd.ncats.io:7474/browser/ .
Limited knowledge and unclear underlying biology of many rare diseases pose significant challenges to patients, clinicians, and scientists. To address these challenges, there is an urgent need to inspire and encourage scientists to propose and pursue innovative research studies that aim to uncover the genetic and molecular causes of more rare diseases and ultimately to identify effective therapeutic solutions. A clear understanding of current research efforts, knowledge/research gaps, and funding patterns as scientific evidence is crucial to systematically accelerate the pace of research discovery in rare diseases, which is an overarching goal of this study.
To semantically represent NIH funding data for rare diseases and advance its use of effectively promoting rare disease research, we identified NIH funded projects for rare diseases by mapping GARD diseases to the project based on project titles; subsequently we presented and managed those identified projects in a knowledge graph using Neo4j software, hosted at NCATS, based on a pre-defined data model that captures semantics among the data. With this developed knowledge graph, we were able to perform several case studies to demonstrate scientific evidence generation for supporting rare disease research discovery.
Of 5001 rare diseases belonging to 32 distinct disease categories, we identified 1294 diseases that are mapped to 45,647 distinct, NIH-funded projects obtained from the NIH ExPORTER by implementing semantic annotation of project titles. To capture semantic relationships presenting amongst mapped research funding data, we defined a data model comprised of seven primary classes and corresponding object and data properties. A Neo4j knowledge graph based on this predefined data model has been developed, and we performed multiple case studies over this knowledge graph to demonstrate its use in directing and promoting rare disease research.
We developed an integrative knowledge graph with rare disease funding data and demonstrated its use as a source from where we can effectively identify and generate scientific evidence to support rare disease research. With the success of this preliminary study, we plan to implement advanced computational approaches for analyzing more funding related data, e.g., project abstracts and PubMed article abstracts, and linking to other types of biomedical data to perform more sophisticated research gap analysis and identify opportunities for future research in rare diseases.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13023-021-02120-9.
A rare disease is defined as any disease that affects fewer than 200,000 individuals in the United States. There are an estimated 25–30 million Americans that are affected by one of approximately 7000 different rare diseases, most of which are poorly understood with unclear underlying biological mechanisms. This knowledge gap leads to challenges for patients, clinicians, and investigators. Patients affected by a rare disease experience delays in diagnosis, as well as a lack of available treatments, clinicians often have limited clinical knowledge and experience impedes their clinical decision making, and investigators struggle with limited patient data and sparse funding for research across most rare diseases [ 1 ]. To help address these challenges, we proposed a detailed analysis of research funding data to (1) enhance understanding of the current funding situation and potential funding opportunities in rare diseases, and (2) identify gaps among current research activities in rare diseases that may be primed for new research.
Compared to common diseases that are more highly prevalent in the population, such as depression or heart disease, research funding is often scarce for rare diseases, in part due to the relatively small number of people affected, and lower prioritization of funding based on the perceived burden of the disease [ 2 ]. In fiscal year 2019, the National Institutes of Health (NIH) were appropriated $39 billion [ 3 ], of which only $38 million (0.1%) was awarded to study a wide range of rare diseases [ 4 ]. Similarly, in the UK, less than 1% of the annual funding budget from three out of the top four UK funders were directed towards rare disease research [ 5 ]. A lack of funding results in less research aiming to understand disease etiology, identify biomarkers for disease diagnosis, develop of novel medications and associated clinical trials, and ultimately, absent treatment options. To solicit and attract more funding for innovative research in rare diseases, there is an urgent need to better understand and assess the current funding situation/trend and address gaps found to persist from retrospectively tracing funding history.
To review the funding landscape, Stehr et al. examined funding circumstances for Batten Disease, a group of rare nervous system disorders, by extracting funding information from publications. Interestingly they discovered 193 funding agencies had supported Batten Disease research to date, which might encourage researchers to continue their pursuits and expand their studies, moving key findings from discovery to application phases [ 6 ]. Franceschi et al. led a study to characterize recent NIH funding for diagnostic radiology departments at US medical schools [ 7 ]. To inform decisions on research direction, Ma et al. examined over 43,000 scientific projects funded over the past three decades and established collaboration networks that revealed major ramifications on future research strategy and government policy [ 8 ]. Packalen et al. performed an analysis on a comprehensive corpus of published biomedical research articles, and found that edge science framed with novel basic science ideas is more often funded by the NIH than less novel science [ 9 ]. These studies either limited their review of the funding landscape to a few specific diseases or diagnostic protocols, or assessed research direction globally. In our study, we aimed to develop an approach to systematically overview funding trend across different disease categories or individual diseases, and identify research gaps globally or locally (i.e., for individual disease category or disease) to further support rare disease research.
To overcome the challenge of handling large amounts of data accumulated given scientific advancements, knowledge graphs have attracted a lot of interest in the biomedical domain, as they can be leveraged to semantically represent relationships among large-scale data [ 10 ]. Ding, et al. constructed a PubMed knowledge graph (PKG) by extracting bio-entities from 29 million PubMed abstracts with integrated funding data through the NIH ExPORTER, in order to measure scholarly impact, knowledge usage and transfer, and profile authors and organizations based on their connections with bio-entities [ 11 ]. In our previous work, we developed an integrative knowledge graph in Neo4j [ 12 ], named NCATS GARD Knowledge Graph (NGKG) that contained a large volume of biomedical and research data pertinent to rare diseases [ 13 ]. Inspired by these published studies, we proposed to aggregate and represent funding data for rare diseases in a semantic manner as a knowledge graph. While the PKG incorporates publications and funding to gain insights and approaches to connect researchers with common research interests, our primary goal is to not only assess the funding landscape of rare diseases, but also identify research status and knowledge gaps, which can be applied to promote novel research for rare diseases.
Rare disease resources
In this study, we incorporated rare disease information from the Genetic and Rare Diseases (GARD) [ 14 ] and Monarch Disease Ontology (MONDO) [ 15 ]. GARD is a public health information center managed by the Office of Rare Diseases Research (ORDR) within the National Center for Advancing Translational Sciences (NCATS), and retains curated disease information for about 7000 rare diseases. MONDO is a semi-automatically constructed ontology that merges multiple disease resources to yield a single coherent merged ontology [ 15 ]. We accessed GARD and MONDO from our previously developed NCATS GARD Knowledge Graph (NGKG) in Neo4j [ 13 , 16 ].
NIH funding resource
Research Portfolio Online Reporting Tools Expenditures and Results (RePORTER) is a key component of the NIH’s “open government” initiative to provide more transparency into NIH activities, including information on NIH expenditures and the results of NIH supported research [ 17 ]. ExPORTER provides bulk administrative data found in RePORTER to the public for detailed analyses or to load into their own data systems [ 18 ]. ExPORTER provides downloadable versions of the data accessed through the RePORTER interface, and includes information about projects, publications, patents, and clinical studies. In this study, we downloaded and cleaned the funded projects and associated publications from ExPORTER, and stored the data in a MySQL database, from where we obtained data for the analysis described in this study.
Rare disease data preparation
We extracted 6305 GARD rare diseases from the NGKG in Neo4j [ 13 , 16 ]. In order to adopt disease categories from MONDO to organize these extracted rare diseases, we only included 5001 GARD rare diseases that have one-to-one exact mapping to MONDO based on one MONDO property, “MONDO:equivalentTo”. As a proof-of-concept with minimized manual validation, we did not include one-to-many, many-to-many or any other partial mappings between GARD and MONDO concepts, which will be included in a future study.
To enable review and analysis of research funding by disease categories, we mapped GARD diseases to MONDO disease categories. MONDO contains three main branches in its disease classification tree, namely, “Disease Characteristic”, “Disease or Disorder” and “Disease Susceptibility”. In this study, we focused on the branch of “Disease or Disorder” (MONDO: 0000001) in MONDO obo file [ 19 ], from where we extracted 32 root disease categories, including congenital abnormality, acute disease, disorder involving pain, serpinopathy, psychiatric disorder, visceral myopathy, and post-infectious disorder, etc. (A complete list of 32 root disease categories can be found in Additional file 1 ) We mapped those 5001 GARD diseases to the 32 root categories accordingly by iteratively searching the MONDO disease hieratical tree. It is worthy to note that most GARD diseases map to more than one MONDO disease categories.
NIH funded project identification for rare diseases
NIH ExPORTER provides detailed information about funded projects, including project titles and project abstracts in free text. We assumed that if the disease name is mentioned in the project title, it is likely that the project was proposed to conduct research investigation on this disease. Hence, we proposed to map GARD disease names to project titles, to identify a list of funded projects for each individual GARD disease via two steps as described below. Before mapping, we excluded projects with invalid project titles, such as, “13.358”, “CFDA NO. 13-299”.
Mapping based on name match
We executed ‘LIKE’ SQL operator as exact string match to identify projects with disease names mentioned in the project titles from the MySQL database, which stores the cleaned funding data. To avoid any mis-mappings, we applied not only full disease names but also alternative disease names in the ‘LIKE’ operator. Since abbreviations occurring in disease names and/or synonyms might cause incorrect mappings, disease names and synonyms with less than 4 characters were excluded for mapping.
Mapping based on semantic annotation.
As a complementary step, we semantically annotated project titles in free text by using MetaMap annotator [ 20 ], and then mapped GARD diseases to those generated annotations. MetaMap annotator produces a list of annotations in the Unified Medical Language System (UMLS), including UMLS semantic types, its preferred names, etc. Figure Figure1 1 shows a snapshot of annotation results generated for a project entitled “The Natural History of Mucolipidosis Type IV”. To avoid any incorrect mappings, we excluded those annotated concepts with less than 4 characters for further analysis, for example: “RNS (UMLS: C1850106)” as one annotation for the project title of “HAMPTON INSTITUTE'S CONTINUING EDUCATION PROGRAM FOR RNS”.

Annotation results generated by MetaMap (The fields in bold were extracted and applied to map to GARD diseases, “C0238386” is UMLS identifier, “Mucolipidosis Type IV” is UMLS preferred name, “[dsyn]” is one UMLS semantic type, “Disease or Syndrome”)
In order to establish the mappings between funded projects and GARD diseases, we mapped annotations generated from each project title to GARD diseases based on UMLS mappings since MetaMap output is in the UMLS, as shown in Fig. Fig.1. 1 . We retrieved mappings between GARD diseases and the UMLS via two steps. First, we obtained the UMLS mappings that were curated by GARD. Next, we obtained the UMLS mapping from MONDO for the GARD disease if the GARD disease concept was exactly matched to the MONDO concept and this MONDO concept had an external mapping to the UMLS. For instance, Bloom syndrome (GARD:0000915) is exactly matched to one MONDO concept of Bloom syndrome (MONDO:0008876) that is mapped to one UMLS concept (UMLS:C0005859). Thus, with UMLS:C0005859, we were able to map this GARD disease to one project entitled “BLOOM'S SYNDROME--DNA LIGASE AND IMMUNODEFICIENCY” with one annotation of Bloom Syndrome (UMLS:C0005859).
Once we established the connections between GARD diseases and funded projects through the above steps, we designed a data model to semantically capture and represent different types of data extracted from those funded projects and their associated data, such as, publications or principal investigators.
Primary class definition
We defined seven primary classes, namely, Disease Category, Disease, Funded Project, Funding Agent, Principal Investigator, Publication, and Journal. These classes capture a full spectrum of information present in NIH funding data and enable linkages to other different types of data for directing more sophisticated research on rare diseases, which will be described in the “ Discussion ” section.
Object property definition
To capture semantic relationships among those primary classes, we defined object properties as shown in Table Table1 1 .
Defined object properties
Data property definition
We defined a list of data properties shown in Table Table2 2 to link data values for each individual concept.
Defined data properties
a MONDO ID was used as category_id because MONDO disease categories were adopted
NIH funding knowledge graph
Based on the data model we described above, we loaded the mapped funding data to a knowledge graph hosted in Neo4j. To be specific, different types of data have been loaded and represented with those seven primary classes as nodes accordingly; object properties were applied to establish semantic connections between different nodes as edges, and data properties were attached to corresponding nodes as node properties. The knowledge graph is publicly assessable without login requirement at http://grants4rd.ncats.io:7474/browser/ .
Results of rare disease data preparation
A total of 5001 unique GARD rare diseases were categorized based on the MONDO disease classes. Table Table3 3 shows the categorization results. Only 799 GARD diseases belonged to a single MONDO category, while most GARD diseases were mapped to multiple MONDO disease categories. For example, GARD:0006735 (Hypophosphatemic rickets) is mapped to three different MONDO disease categories: MONDO:0003847 (Mendelian disease), MONDO:0003900 (connective tissue disease), and MONDO:0021199 (disease by anatomical system). There are 52 GARD diseases that were not grouped into any of MONDO disease categories, because they were either mapped to obsolete MONDO diseases or another two MONDO disease category branches, “Disease Characteristic” and “Disease Susceptibility”, which were excluded from this study.
Results of GARD diseases to MONDO disease categories
Results of NIH funding data mapping
Results of nih funding data retrieval.
We downloaded the funding data with funding year spanning from 1985 to 2019 from NIH ExPORTER [ 18 ]. A total of 2,457,303 distinct applications with 654,347 unique project titles, 886,895 unique project abstracts, and 2,555,300 publications are cleaned and stored in a MySQL database.
Results of NIH funding data mapped to GARD
Two types of mapping results have been generated via exact name (i.e., String) match and MetaMap annotations.
Disease name mapping results
Both GARD disease names and synonyms have been applied to map project titles. 1104 GARD diseases were mapped to 21,027 project titles, which correspond to 63,692 NIH funded applications. Since one project can be funded for multiple years as multiple applications with the same project title, the number of mapped applications is larger than the number of project titles.
MetaMap annotation results
652,975 unique project titles were annotated by the MetaMap annotator and 5,039,735 annotations were generated. To map GARD diseases to those annotations based on UMLS mappings, we first retrieved UMLS mappings for GARD diseases from GARD and MONDO. Specifically, 3468 GARD diseases with UMLS mappings were extracted from GARD, and out of 15,629 MONDO concepts with UMLS mappings, 3980 MONDO entries were mapped to 4032 GARD diseases and corresponding UMLS mappings, which were assigned to those GARD diseases. Together, 1146 GARD diseases were mapped to 13,695 project titles via UMLS mappings.
By merging mapping results from the above two steps, 1294 GARD diseases were successfully mapped to 72,577 funded applications, which corresponds to 45,647 distinct projects.
Statistical results of NIH rare disease funding data in the Neo4j knowledge graph
Summarized statistical results of individual concepts belonging to each primary class are shown in Fig. Fig.2 2 .

Statistics of rare disease funding Neo4j knowledge graph
Case studies
To demonstrate the use of this integrated knowledge graph with rare disease funding data, we performed two types of case studies, (1) funding landscape assessments for an overview of the current funding landscape; (2) evidence-based research opportunity identification for supporting research in rare diseases. Cypher Queries have been composed and executed to perform case studies described in this section, and more details about those Cypher Queries are included in the Additional file 2 .
Current funding landscape
To review NIH funding scenario in rare diseases, we composed several queries using Cypher, which is Neo4j’s graph query language that allows users to store and retrieve data from the graph database [ 21 ], to search against funding data in our Neo4j. Cypher Query 1 was constructed to access funding circumstances by disease categories and the result is listed in Table Table4. 4 . Since one disease might be grouped into multiple disease categories, duplicates occurred when the number of GARD diseases, number of projects and total cost by each category were summed. Regardless, the numbers listed in Table Table4 4 consistently reflect funding priorities with the consideration of disease burden applied by NIH. In addition, we retrieved funding amounts in the last twenty years for the top five most funded disease category, which is shown in Fig. Fig.3. 3 . It is worthy to note that no funding cost data is available before the year of 2000 in our downloaded data. From Fig. Fig.3, 3 , besides infectious diseases, big jumps were present due to the Ebola and Zika outbreaks that occurred during that time frame, the increasing funding trends are observed for other categories.
Funding landscape in rare diseases by disease categories

Funding trend by year for the top five most funded disease categories
Cypher Query 1
MATCH p = (n:Project)<-[:isInvestigatedBy]-(d:Disease)-[:isClassOf]-(c:DiseaseCategory) RETURN c.name AS Disease_Category, COUNT(DISTINCT d.gard_id) AS NumOfGARD, COUNT(n.application_id) AS NumOfProjects, SUM(TOINTEGER(n.total_cost)) AS Total_Funding_Amount ORDER BY Total_Funding_Amount DESC
We also evaluated the top 10 funded individual diseases (via Cypher Query 2). The top four funded diseases shown in Table Table5 5 are infectious diseases. Poliomyelitis is considered a global public health emergency and the U.S. is a partner of the Global Polio Eradication Initiative [ 22 ]. Particularly, the CDC and the U.S. Agency for International Development (USAID) work to eradicate polio and have signed onto the Polio Endgame Strategy 2019–2023 [ 23 ]. About half of the world population is at risk for Malaria and the U.S. is the largest donor to the Global Fund to Fight AIDS, Tuberculosis, and Malaria (Global Fund) [ 24 ]. A large amount of research funds are being distributed on research for effective vaccines for Anthrax and Measles. In comparison to other rare diseases, the rest of diseases listed in Table Table5, 5 , such as sickle cell disease with about 100,000 affected people in the US, both of cystic fibrosis and Huntington disease with more than 30,000 affected people in the US, are diseases that are closer to finding a cure or effective treatment, in part because they receive more research funding.
Funding landscape in rare diseases by individual diseases
Cyper Query 2
MATCH p = (d:Disease)-[:isInvestigatedBy]-(n:Project) RETURN d.gard_id AS GARD_ID, d.name AS GARD_Name, SUM(toInteger(n.total_cost)) AS Total_Funding_Amount ORDER BY total_cost DESC LIMIT 10
Evidence-based rare disease research discovery
We aggregated funded projects and their publications into an integrated knowledge graph in Neo4j, which offers opportunities to programmatically support new research for rare diseases.
Research landscape assessment
Funded projects along with their publications show snapshots of their research goals and outcomes, which provides an opportunity to systematically assess the current research status and gaps, and consequently direct future areas for investigation. We enumerated two types of assessment for Neuronal ceroid lipofuscinosis (GARD: 0010739) and Duchenne Muscular Dystrophy (GARD:0007922) as examples.
Research status assessment for neuronal ceroid lipofuscinosis (NCL)
NCLs are classified by their causal gene of CLN (ceroid lipofuscinosis, neuronal), which is given a different number designation as its subtype. Signs and symptoms range in severity and progress at different rates given different gene mutations. The disorders generally include a combination of vision loss, epilepsy, and dementia. Some forms of the NCLs are: CLN1 disease, infantile onset; CLN2 disease, later-onset and so on [ 25 ]. Scientific investigations have been performed for each subtype of NCL (via Cypher Query 3). By manually examining the funded projects and their published studies for NCL, we grouped them based on their studied NCL forms (Fig. (Fig.4). 4 ). Noticeably, these funded projects aim to better understand the molecular mechanism of NCL and discover therapeutic solutions. In particular, several subtypes (CLN2, CLN3 and CLN6) of NCL are more extensively studied than others. There is an Food and Drug Administration (FDA) approved enzyme replacement therapy for CLN2 disease (TTP1 deficiency) called cerliponase alfa (Brineura®) that has been shown to slow or halt the progression of symptoms [ 26 ]. With the exception of CLN2, which is highlighted as “CLN2” in Fig. Fig.4, 4 , there are no approved treatments that can slow or stop disease progression for other forms of NCL disorder. As NCL affects the brain and nervous system, treatments must reach the brain to be effective, but getting the proper enzyme to cross the blood brain barrier can be difficult. For this reason, enzyme replacement therapy can only be used in NCL forms where the affected enzyme is soluble. This includes the subtypes known as CLN1, CLN2, CLN5 and CLN10 [ 27 ]. Thus, learning from the success of Brineura for CLN2 enhances the necessary scientific evidence and understanding of what existing research has been performed in other forms of NCL, and may better inform the direction of further research investigation on NCL.

Research landscape of Neuronal ceroid lipofuscinosis (Light blue nodes denote diseases, orange nodes denote funded projects, magenta nodes denote publications, Dark blue nodes denote principal investigators. CLN1, CLN2, CLN3, CLN4, CLN6 and ACLN are different forms of NCL)
Cypher Query 3
MATCH p = (d:Disease)-[:isInvestigatedBy]->(n:Project)<-[:hasFundedProjectOf]-(m:PrincipalInvestigator) WHERE d.gard_id = 'GARD:0010739' RETURN p
Research study clustering for Duchenne Muscular Dystrophy
Publications introduce novel approaches and/or findings that were proposed and generated from the funded projects. For example, a project entitled “LOCALIZATION OF X-LINKED HYPOPHOSPHATEMIC RICKETS GENE” (APPLICATION ID = “3087091”), has reported two publications, “Mutational analysis and genotype-phenotype correlation of the PHEX gene in X-linked hypophosphatemic rickets.” (PMID: 11502829) and “Mutational analysis of the PEX gene in patients with X-linked hypophosphatemic rickets.” (PMID: 9106524) These two publications specifically reported their investigation on genes for X-linked hypophosphatemic rickets. For a given disease, we clustered research studies by a project or a list of projects with similar research topics, which allows tracking research trajectory, identifying research gaps, and preparing necessary training data for future study. In this case study, we executed Cypher Query 4 to cluster publications associated with the funded projects for Duchenne Muscular Dystrophy (GARD:0006291), a lethal muscle wasting disease caused by the lack of dystrophin, which eventually leads to apoptosis of muscle cells and impaired muscle contractility. Four projects, along with their publications, are shown in Fig. Fig.5. 5 . Based on the project titles shown in gray boxes in Fig. Fig.5, 5 , we deduced that their objectives are all tied to breaching the major barriers to successful therapeutic solutions for Duchenne Muscular Dystrophy respectively via iPSCs, Cas9 and Cx43.

Publications clustered by funded projects for Duchenne Muscular Dystrophy (Blue nodes denote diseases, orange nodes denote funded projects, magenta nodes denote publications. Project titles are shown in the gray boxes)
Cypher Query 4
MATCH p =(d:Disease)-[:isInvestigatedBy]-(n:Project)-[:hasPublication]-(m:Publication) WHERE d.gard_id = 'GARD:0006291' RETURN p
Identify new research opportunities
In this case study, we proposed to gain research insights systematically by analyzing project titles. For instance, given one project entitled “USE OF LEUPROLIDE ACETATE FOR TREATMENT OF PRECOCIOUS PUBERTY”, we know this project was studying leuprolide acetate (Drug) for precocious puberty [ 28 ]. With the disease annotations generated for project titles (described in the “ Identify new research opportunities ” section), we were able to identify potential disease associations via projects, and furthermore to identify new avenues for research. Fig. Fig.6 6 demonstrates an example of looking for potentially relevant diseases to Measles (GARD:0003434) via Cypher Query 5. Obviously multiple rare infectious diseases, including Poliomyelitis, Rubella, Congenital rubella, and Malaria are grouped together, and interestingly Measles and Glioblastoma is linked via one project entitled “Measles Virotherapy for Glioblastoma Multiforme”, highlighted in the red circle shown in Fig. Fig.6. 6 . According to one recently published review paper written by Zhang et al. [ 29 ], “advances and potential pitfalls of oncolytic viruses expressing immunomodulatory transgene therapy for malignant gliomas”, the authors emphasized that the therapeutic efficacy of oncolytic viruses alone is limited, which might motivate novel research to better understand and assess the therapeutic efficacy. Five related publications produced by this project provide additional information to direct future investigation.

Identifying possible research opportunities for Measles via funded projects (Orange nodes denote funded projects, blue nodes denote rare diseases, magenta nodes denote publications)
Cypher Query 5
MATCH p =(d:Disease)-[:isInvestigatedBy]-(n:Project)-[:isInvestigatedBy]-(d1:Disease) WHERE d.gard_id = 'GARD:0003434' RETURN p
In this study, we extracted NIH funding data for rare diseases and semantically represented it as a knowledge graph, which provides an effective way to not only assess the funding landscape in rare diseases, and systematically identify new research gaps/opportunities in rare diseases, but also share identified scientific findings/evidence with funding agencies for recommending their research fund distribution in rare diseases.
Scientific observations from data preparation
Given various representations of disease names applied in the project titles, to accurately map diseases to projects via project titles and avoid any mis-mapping, we applied a complementary mapping strategy using string match and semantic annotation. For instance, one project entitled “Discovering Novel Treatments for Batten Disease” could only be mapped to “Batten Disease (GARD:0010739)” via string match based on the disease name, instead of semantic annotation. The MetaMap annotations generated from the project title is “Juvenile Neuronal Ceroid Lipofuscinosis (UMLS: C0751383)”, which is different from the UMLS mapping (UMLS:C0027877) associated with this GARD disease. Another project entitled “TOXIC METALS, MEMBRANE SIGNALING, AND CELL GROWTH” is mapped to “Heavy metal poisoning (GARD:0006577)” via semantic annotation instead of string match, since the MetaMap annotator generates one annotation of “toxic metals (UMLS: C0274869)”, which is mapped to the UMLS associated with that GARD disease. Clearly, even though we applied this complementary strategy to establish mappings between projects and diseases, we may miss relevant projects with the disease mentioned in the project abstract, instead of in the project title. To avoid any missing data, several additional steps have been proposed and described in the “ Future work ” section.
Scientific insights derived from the case studies
We performed two types of case studies, funding landscape and research landscape analysis respectively to demonstrate the use of our funding knowledge graph for evidence-based rare disease research discovery.
Funding landscape assessment
Accessing funding data allows researchers to examine funding situations and funding opportunities in rare diseases. Our case studies illustrate that the funding trend consistently reflects the disease burden to the public health, e.g., infectious diseases received the most amount of funding support over years. A big jump regarding the total funding amount for infectious diseases between 2013 and 2017, coincides with the Ebola and Zika outbreaks that occurred during that timeframe. Another inspiring message derived from our analysis is that the amount of funding for supporting rare disease research has been increasing yearly, which might encourage and motivate more scientists to devote more of their research to rare diseases.
Scientific evidence-based rare disease research discovery
Analysis of funding data not only provides a systematic way to outline a complete research spectrum for an interested disease (category), but also generates scientific evidence supporting rare disease research discovery. For instance, the case study of Neuronal Ceroid Lipofuscinosis (NCL) illustrates that several different types of NCL have been investigated intensively and one therapeutic success on one type of NCL, namely, CLN2, which will enable us to dive into it and derive insight and knowledge about the status and gaps of research on NCL in further investigation. In addition, we demonstrated the power of aligning funded projects with their publications to direct research. As shown in the case study of Duchenne Muscular Dystrophy, it becomes feasible to cluster research papers based on a funded project or a list of projects with a similar research topic, which enables researchers to systematically track research pathway and programmatically prepare training data for future study. In this study, publication clusters were solely based on single projects; we propose to cluster publications with multiple projects with similar research topics by implementing Natural Language Processing (NLP) algorithms to analyze project abstracts.
Limitations and future work
By reviewing the MONDO categories applied to organize rare diseases, some categories, for instance, “Mendelian disease”, “disease by anatomical system”, are broad categories consisted of many individual diseases; In addition, as shown in Table Table3, 3 , most rare diseases are grouped into multiple MONDO categories. We propose to extend our disease category mapping to a higher granularity level as the next step, to precisely reflect funding distribution and assess research landscape by disease categories.
In this study, mappings between rare diseases and funded projects were established based on project titles. To avoid any mis-mappings with rare diseases that are mentioned in the project abstracts instead of project titles, we propose to expand our analysis with project abstracts. In addition, analyzing funding data from other resources, such as “funding” or “Acknowledgements” sections included in the publications to introduce the funding sources supporting their research, is another proposed extension task. Although the aforementioned extensions have been proposed as future work by aligning with our current project plan, we anticipated that we will still miss some data, such as, (1) projects without project titles and/or abstracts, (2) non-US government funded projects; (3) projects with only a single funding source (not all) was acknowledged in the publications. These are beyond the scope of this project and will be planned in the future study.
Insights and lessons drawn from the case studies were heavily relied on manual interpretation in this preliminary study, and we propose to transform currently manual processes into automated processes by implementing NLP and advanced machine learning techniques. Given the nature of rare diseases, limited data and knowledge about those diseases compared to common diseases, collaborative efforts seem more important and critical. We propose to discover methods to better promote research collaboration by connecting investigators with the requisite expertise and shared research interest from our funding knowledge graph. To that end, scientists can work collaboratively to pool patients, data, experience and resources together to support more innovative research in rare diseases. We are also interested in investigating the relationship between medical cost using information from datasets such as the AHRQ’s Healthcare Cost and Utilization Project (HCUP) [ 30 ] and research cost (i.e., NIH funding) in rare diseases, with the hypothesis that higher research funding should lead to improvements in earlier diagnosis of a rare disease and lower medical costs. Identifying overlaps and discrepancies between research status and medical situation will guide both further investigation and better inform decision makers on how to stimulate and advance research across more rare diseases.
In this preliminary study, we developed an integrative knowledge graph to semantically represent NIH research funding data for rare diseases and successfully demonstrated its use of directing and promoting scientific evidence based rare disease research discovery. With the success and lessons learned from this study, we propose multiple improvements/extensions as the next step, to fully utilize funding data to accelerate the pace of rare disease research.
Abbreviations
Authors' contributions.
The work was conceived by QZ, who designed and performed data analysis and drafted the manuscript; TN helped on Neo4j development; GA helped on case studies with funding landscape assessment; ES, EAM helped on case studies with research landscape assessment and manuscript edition; TS, YX, JD and AP participated in the project discussion and edited the manuscript. All authors read and approved the final manuscript.
Open Access funding provided by the National Institutes of Health (NIH). This research was supported by the Intramural and Extramural research program of the NCATS, NIH.
Availability of data and materials
Declarations.
Not applicable.
None declared.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Genomics-focused biomedical research secures $6.5 million to address complex health challenges

Genomics-focused biomedical research breakthroughs in cancer, inherited eye disease and common gynaecological disorders are set to be accelerated by a $6.5 million funding boost.
The Advanced Genomics Collaboration (TAGC) has awarded the funding to four new Innovation Projects led by University of Melbourne researchers collaborating with partners across the Melbourne Biomedical Precinct.
The four projects will also gain access to powerful DNA sequencing technology in TAGC’s world-leading genomics hub made possible through a partnership between the University and global biotech company Illumina , with support from Invest Victoria .
The four projects:
- Associate Professor Lauren Ayton and Dr Ceecee Britten-Jones from the Department of Optometry and Vision Sciences and the Centre for Eye Research Australia will establish a comprehensive patient dataset of inherited retinal diseases, providing information to drive new treatments and therapies for the most common cause of blindness in working-aged Australians.
- Associate Professor Richard Tothill from the Centre for Cancer Research is partnering with Professor Sarah-Jane Dawson and clinicians and researchers at the Peter MacCallum Cancer Centre to develop a rapid multi-omic liquid biopsy blood test using whole genome sequencing to resolve the difficult diagnosis of cancer of unknown primary origin.
- Professor Peter Rogers at the Department of Obstetrics and Gynaecology will perform next generation sequencing to find new solutions for patients with common endometrial related health problems such as abnormal menstrual bleeding, infertility and pregnancy disorders at The Women’s and Mercy Hospital.
- Professor Niall Corcoran from the Department of Surgery at the Royal Melbourne Hospital will lead a study using genomic testing and predictive disease modelling to guide therapy and reduce the burden of prostate cancer diagnosis, which evidence suggests is over-diagnosed and over-treated in early disease.
Dean of the Faculty of Medicine, Dentistry and Health Sciences Professor Jane Gunn congratulated the projects.
“The TAGC Innovation Projects are a great example of industry and research coming together to advance the translation of genomic medicine,” Professor Gunn said.
“Access to new generation sequencing technology will allow researchers to unlock information embedded in our DNA and develop targeted solutions to complex health challenges.
“These exciting new projects will provide deeper insight into common diseases and health problems impacting many Australians, allowing them to benefit from the life-changing potential of genomics informed precision health care.”
Commercial Lead Oceania, Illumina Simon Giuliano said: “As a genomics pioneer, we have shown through focused and continued innovation how unlocking the power of the genome can exponentially improve the human condition.
“Congratulations to the recipients of the innovation project grants. It is great to see researchers being enabled through the TAGC, to use the power of next generation sequencing to explore new treatments and therapies in conditions and diseases such as inherited blindness, gynecological disorders and cancer.”
19 Mar 2024
Media contact.
Find an Expert
Related news
Australian institute for infectious disease announces early works contractor, societal shift necessary to support women through menopause, experts say.
- International edition
- Australia edition
- Europe edition

She beat a rare liver cancer – and now works with her father to find more cures
Cancer scientist decides to study the tumour that once afflicted his small daughter – and now her work is adding to his project’s success
Elana Simon was 10 years old when she started to experience severe pains in her abdomen. For two years, puzzled doctors put forward diagnoses including lactose intolerance, Crohn’s disease and stress. It was not until 2008 that they pinpointed the real cause. Elana was suffering from fibrolamellar carcinoma (FLC), a rare, usually lethal, form of liver cancer.
“In a way, it was comforting to have a word for what was wrong with me after so much confusion about my condition,” Elana told the Observer . “Pre-diagnosis, my life was a mixture of discomfort and fear. Now I had something to focus on.”
Surgeons were able to cut out the tumour, which had yet to spread out of her liver, and Elana survived. Indeed, she has triumphed over her affliction in a remarkable manner. After studying at Harvard, she joined the laboratory at New York’s Rockefeller University run by her father, Prof Sanford Simon, who since his daughter’s diagnosis has dedicated his career to tacking FLC.
“The moment I learned of Elana’s condition is seared into my memory,” Prof Simon told the Observer . “I was determined to find out what caused it and find ways to counter it.
“I was working as a cell biologist at the time and could have cruised through the rest of my career. Elana’s diagnosis changed everything and I completely altered the direction of my research, which is now focused entirely on the cancer.”
Research at Prof Simon’s laboratory has since led to several major breakthroughs in pinpointing the cause and behaviour of FLC. Now that work has been further boosted by a £20m grant from Cancer Research UK, given this month as part of its Cancer Grand Challenges awards. The grants are awarded – in partnership with the National Cancer Institute in the US – to research groups whose novel ideas offer the greatest potential to advance cancer research.
A consortium that includes Prof Simon’s laboratory has been given its Cancer Grand Challenge grant to help it develop new, easily accessible treatments for conditions such as FLC. At present, treatments for solid tumours in children still rely on decades-old chemotherapy regimes, and the aim of the programme is to develop new, less-complicated interventions.
Their win says much about the success scientists have had with FLC in recent years. When Prof Simon decided to start studying the cancer, its cause and roots were still a puzzle. Was it a single disease or were cases made up of people with different conditions that all had similar symptoms.
In addition, the cause of the disease was a mystery. “We didn’t know whether it was inherited, or caused by a specific environmental insult,” he said.
It was then that Elana – who at the time was a high school student, but is now studying for a PhD in computer science and cancer biology at Stanford University in California – stepped in. “I had ended up making a lot of friends who were about my age and who were going through various paediatric cancer diagnoses and treatments,” she said. Elana and one of these friends launched a YouTube appeal for other people with that cancer to submit tissue samples to her father’s laboratory. They ended up with 15 samples in total, and these played a crucial role in helping the team at Rockefeller make key discoveries about the disease.
“First, we found that it was a single disease with one mutation that was the same for each tumour sample. This was causing the disease,” said Elana who had pioneered this part of the project.
In addition, the team found that FLC was not an inherited condition. “It is caused by a single environmental insult, and that was crucial,” added Prof Simon, “for it suggested that if you cut out a tumour before it had spread, that meant you could halt the disease. It was unlikely to re-occur.”
Fibrolamellar carcinoma was very rare, Prof Simon acknowledged. Some studies suggest there are only a few hundred cases worldwide each year, though more recent analysis suggests the figure may reach thousands.
“The crucial point is that if you look at common cancers in adults, you come across all sorts of background genetic mutations in cells, and that makes it difficult to assess what is driving the disease,” he said. “However, with a childhood cancer like FLC, relatively few mutations develop and so it becomes much easier to understand what is going on. In this way, cancers like these provide critical insights that reveal general principles.”
Another cancer this applied to was the rare retinoblastoma eye tumour, Prof Simon said. Studies of patients with the condition revealed the existence of tumour suppressor genes: these normally limit the spread of cancer cells but occasionally fail. Therapies based on this discovery are now being developed for many cancers.
“That is what we hope we can do with the work we are doing on this carcinoma,” said Elana. “It will not just help people like me who get FLC, but will provide general insights into other cancers. The Cancer Grand Challenge funds will make a big difference to those efforts.”
At the same time, it is hoped that easily administered therapies for FLC will be developed from the insights already gained about the disease.
“Ultimately, we want to create drugs that can be administered as pills in places where the health provision is poor,” said Prof Simon. “It is the ultimate goal.”
- Cancer research
- The Observer
- Medical research
Most viewed
- Staff Contacts
College Resources
- Administration
- Alumni & Giving
- College News
- Publications
- Departments & Units
Contact Information
Scovell Hall Lexington, KY 40546-0064
Search the Martin-Gatton College of Agriculture, Food and Environment
Kentucky tick surveillance project accepting submissions, advancing public health research and safety.
The Kentucky Tick Surveillance Project, housed in the University of Kentucky’s Department of Entomology, offers a unique opportunity for the community to contribute to tickborne disease understanding — furthering public health research in the state.

By Jordan Strickler Published on Mar. 19, 2024
The Kentucky Tick Surveillance Project is now accepting tick-testing submissions from Kentucky residents. This project, produced by the University of Kentucky Martin-Gatton College of Agriculture, Food and Environment, aims to improve knowledge about where ticks are found and the diseases they might carry.
The results help further public health research at the state level and alleviate concerns for citizens worried about tickborne illnesses.
"The Kentucky Tick Surveillance Project directly contributes to our understanding of tickborne diseases and their distribution across the state," said Jonathan Larson, UK Department of Entomology assistant professor. "By the community participating, Kentuckians are not only aiding in crucial public health research but are also taking proactive steps toward safeguarding their own health and their neighbor’s."
To ensure the safety and integrity of the samples, and to comply with postal regulations, participants are urged to follow strict guidelines when preparing ticks for submission:
- Do Not Mail Live Ticks : Ensure that ticks are not alive when sent.
- Avoid Liquid Alcohol : Do not send samples in containers of liquid rubbing alcohol.
- Proper Packaging : Avoid using only paper envelopes for mailing ticks as they may get damaged in mail processing machines.
- Avoid Taping Ticks : Do not place ticks between pieces of tape.
Participants must complete a submission form before sending their tick sample. This form is crucial for processing and can be found at https://bit.ly/49SixpO .
Samples without a completed form will not be accepted. Detailed instructions for preparing the tick sample, including necessary supplies and packaging steps, ensure samples arrive in good condition for testing.
Important Considerations:
- Selective Testing : Ticks will not be tested for all possible pathogens. The project focuses on gathering data for surveillance purposes.
- Notification Process : Submitters will only be contacted if their submissions test positive for pathogens. There is no notification of negative results.
- Backlog and Time : Due to the project's volunteer nature, there is a testing backlog. Participants should expect delays.
- Not a Substitute for Medical Care : If a tick has bitten you and has symptoms, seek immediate medical attention. Do not rely solely on submitting the tick for health decisions.
"Community involvement is the backbone of the Kentucky Tick Surveillance Project," Larson said. "Every tick submitted is a piece of the puzzle in understanding our state's tickborne disease landscape. We thank everyone who participates for their contribution to this important work."
Please send all ticks to the address below:
Tick Surveillance Program C/O Subba Palli Department of Entomology S-225 Ag Science Center North Lexington, KY, 40546-0091
For more information on the Kentucky Tick Surveillance, visit https://entomology.ca.uky.edu/ticksurveillance2022 .
The Martin-Gatton College of Agriculture, Food and Environment is an Equal Opportunity Organization with respect to education and employment and authorization to provide research, education information and other services only to individuals and institutions that function without regard to economic or social status and will not discriminate on the basis of race, color, ethnic origin, national origin, creed, religion, political belief, sex, sexual orientation, gender identity, gender expression, pregnancy, marital status, genetic information, age, veteran status, physical or mental disability or reprisal or retaliation for prior civil rights activity.
Contact: Jonathan Larson, [email protected] Media Requests: C.E. Huffman, [email protected]
Have a news, success story, or PR idea?
Many of the stories we cover start as suggestions from our partners in the college and across Kentucky. If you have an idea, please share it with us by clicking below.
Related News
Kentucky high school crop scouting competition registration now open.
By Jordan Strickler Published on Mar. 20, 2024
Eastern Tent Caterpillars make their 2024 Kentucky debut
By Jordan Strickler Published on Mar. 8, 2024
UK Extension agent honored with Grain Crops Science Service Award, recognizing decades of dedication
By Jordan Strickler Published on Feb. 28, 2024
- Departments
- Partnerships
- Alumni + Giving
- Dean — Nancy Cox, Ph.D.
- S123 Ag. Science Center North
- Lexington, KY 40546-0091
- Accreditation
- An Equal Opportunity University
- Report a Site Issue
© 2024 University of Kentucky, Martin-Gatton College of Agriculture, Food and Environment
UTA federal research projects add $38M to economy
Tuesday, Mar 19, 2024 • Katherine Egan Bennett : contact
.ashx?revision=7960ab7b-e061-4d85-b6c7-472ba813b6b5)
The economic impact of federally sponsored research at The University of Texas at Arlington was $38 million in 2022, with expenditures spread among 725 unique vendors, according to a new report. Of that total, the University spent about $24 million on research-related goods and services in Texas.
The research dollars also supported the salaries of 1,562 people during this time—including 517 faculty and 746 students.
“Research at UTA helps solve some of society’s most important problems, and it is also a vital economic driver in our economy and in the careers of our students and researchers,” said Kate Miller, vice president of research and innovation. “We’re proud that UTA’s collaborations with private and public research organizations led to the purchase of new scientific equipment and technology that has such an economic impact on our region.”
According to the report produced by the Institute for Research on Innovation and Science (IRIS), the top federal agencies supporting UTA research are the National Science Foundation, National Institutes of Health, Department of Defense, Department of Transportation and the Department of Commerce.
In the past five years, UT Arlington has purchased research-related goods and services from vendors located in all 38 of Texas’ federal congressional districts.
“Our reports clarify and explain the economic impact of university research through many different lenses,” said IRIS Executive Director Jason Owen-Smith, a professor of sociology and executive director for research analytics at the University of Michigan. “Through these data-driven reports, our goal is to better understand, explain and ultimately improve the public value of higher education and research.”
This report is based on administrative data UTA supplied to IRIS, which was then merged with other public and private datasets. Reports are available to IRIS members. No individual businesses, employees or students are identifiable in the reports.
Nearly 500 researchers from more than 100 institutions have accessed IRIS data through its virtual data enclave, and more than 40 published papers and three books have used the data. IRIS is a national consortium of research universities organized around an institutional review board-approved data repository, housed at the Institute for Social Research at the University of Michigan.

IMAGES
COMMENTS
Participants in clinical studies help current and future generations. Through these studies, researchers develop new diagnostic tests, more effective treatments, and better ways of managing diseases with genetic components. Participants in studies are actively involved in understanding their disorder and current research.
Diseases are abnormal conditions that have a specific set of signs and symptoms. Diseases can have an external cause, such as an infection, or an internal cause, such as autoimmune diseases ...
To semantically represent NIH funding data for rare diseases and advance its use of effectively promoting rare disease research, we identified NIH funded projects for rare diseases by mapping GARD diseases to the project based on project titles; subsequently we presented and managed those identified projects in a knowledge graph using Neo4j ...
Research is strengthening surveillance, rapid diagnostics and development of vaccines and medicines. Public-private partnerships and other innovative mechanisms for research are concentrating on neglected diseases in order to stimulate the development of vaccines, drugs and diagnostics where market forces alone are insufficient.
Developments in Alzheimer's disease research. One of the hallmarks of Alzheimer's is an abnormal buildup of amyloid-beta protein. A study in mice suggests that antibody therapies targeting amyloid-beta protein could be more effective after enhancing the brain's waste drainage system.In another study, irisin, an exercise-induced hormone, was found to improve cognitive performance in mice.
Research Projects. Infectious diseases, particularly emerging pathogens, are an important public health concern across the globe, as well as the institute's home state of South Carolina. ... The Institute for Infectious Disease Translational Research is a multi-primary investigator endeavor, with highly regarded experts in various fields ...
Main Areas of Focus. Research projects will include surveillance studies to identify previously unknown viral causes of febrile illnesses in humans, the animal sources of viral or other disease-causing pathogens, and to determine what genetic or other changes make these pathogens capable of infecting humans.
Our research is focused on identifying molecular mechanisms that allow epidemic S. aureus strains such as USA300 to successfully disseminate. This project uses a combined approach of whole-genome comparative sequencing of longitudinally collected samples, genetic manipulation, and functional studies on bacterial adhesion and survival.
Abstract. As the 21st century unfolds, strategies to prevent and control infectious diseases remain an area of vital interest and concern. The burden of disease, disability, and death caused by infectious diseases is felt around the world in both developed and developing nations. Moreover, the ability of infectious agents to destabilize ...
Introduction. Rare diseases (RDs) are an emerging public health priority. RD refers to a disease that affects a small number of people in a population ().There are 6,000-8,000 unique RDs identified, with approximately 80% being genetic in origin, and 50-75% being pediatric onset (1-3).They are often chronic, progressive, and debilitating, and can lead to significant morbidity and ...
The Division of Infectious Diseases participates in graduate programs and research in microbiology and molecular genetics, immunology and molecular pathogenesis, as well as population biology, ecology, and evolution. Additional opportunities are available for basic science research in: Toxin-mediated infections. Intracellular pathogens. Influenza.
Infectious Disease, Opportunistic Infection. 9/1/14. 5/31/19. GS-US-367-1172: A Phase 3, Global, Multicenter, Randomized, Open-Label Study to Investigate the Safety and Efficacy of Sofosbuvir/Velpatasvir/GS-9857 Fixed-Dose Combination for 8 Weeks Compared to Sofosbuvir/Velpatasvir for 12 Weeks in Direct-Acting.
A College senior's research project has shown a way to more quickly understand the characteristics of emerging diseases, ... Further, the work showed that there was a clear pattern of disease progression, with early symptoms of fever, dry cough, sore throat, and chills being followed by more severe shortness of breath by 5.22 days, matching ...
IN RARE DISEASE RESEARCH PROJECTS BASIC PRE-CLINICAL TRANSLATIONAL & SOCIAL Written by the members of the working group PENREP* Guide first published in July 2020 ... Key concepts of the research project are identified through close consultation with local / national Patient Organisation to ensure objectives of the research
Research Projects. Cardiovascular dysfunction accounts for some of the most common and devastating diseases in the country, as well in the institute's home state of South Carolina. This research promises to uncover new understandings and breakthroughs to reduce mortality and morbidity rates due to cardiovascular disease.
If yes, you will find this list interesting. This category comprises hot topics in infectious disease fields. Consider some of these ideas for your research paper. The virology, epidemiology, and prevention of COVID-19. The diagnosis of COVID-19. Prevention vaccines for SARS-CoV-2 infection.
The United States should fund a new dedicated Disease X Medical Countermeasure Program that leverages technologies and vaccine platforms most suitable to the viral families that are likely to cause future catastrophic disease outbreaks. Medical countermeasures against 1 member of a viral family could easily be adapted to another member quickly ...
A genetic disorder is a disease caused in whole or in part by a change in the DNA sequence away from the normal sequence. Genetic disorders can be caused by a mutation in one gene (monogenic disorder), by mutations in multiple genes (multifactorial inheritance disorder), by a combination of gene mutations and environmental factors, or by damage to chromosomes (changes in the number or ...
The joint DMS/NIGMS initiative also supports research related to infectious diseases that may lead to a more general understanding of the underlying problems and have broader applicability (as opposed to applications to a specific disease). The programs mentioned above focus on individual or small group research projects.
Mayo Clinic research finds a connection between Social Network Index score and AI-determined biological age ROCHESTER, Minn. — A new study from Mayo Clinic finds ...
Science Fair Project Idea. In a survey conducted from 2007 to 2010, the U.S. Centers for Disease Control and Prevention reported that about 49% of people in the United States had taken at least one prescription drug during the past month, and about 22% of people had taken three or more prescription drugs. People are prescribed drugs all the ...
In this project we use a 10-year-old vineyard with 14 wine grape cultivars grown under commercially relevant conditions at a University of California research station to study the progression of Pierce's disease in inoculated vines. We infected grapevines with Xf in May 2021.
The chosen disease for this research project is Alzheimer's disease, a progressive neurodegenerative disorder that primarily affects older adults. Alzheimer's disease is the most common cause of dementia, characterized by a decline in cognitive function and memory loss. It is a significant public health concern, with an increasing prevalence ...
Researchers at the Allen Institute are pursing "moonshot" projects, like mapping the human brain, in hopes of understanding the organ and to help fight brain diseases.
Above: Gottheimer at Cooperman Barnabas Medical Center. LIVINGSTON, NJ — Today, March 19, 2024, U.S. Congressman Josh Gottheimer (NJ-5) announced new legislation, the Securing Equal Access to Research, Care, and Health, or SEARCH Act, to help fund new efforts to increase the number of women in clinical research trials, leading to treatments and cures for rare diseases and blood disorders ...
Methods. To semantically represent NIH funding data for rare diseases and advance its use of effectively promoting rare disease research, we identified NIH funded projects for rare diseases by mapping GARD diseases to the project based on project titles; subsequently we presented and managed those identified projects in a knowledge graph using Neo4j software, hosted at NCATS, based on a pre ...
Genomics-focused biomedical research breakthroughs in cancer, inherited eye disease and common gynaecological disorders are set to be accelerated by a $6.5 million funding boost. The Advanced Genomics Collaboration (TAGC) has awarded the funding to four new Innovation Projects led by University of Melbourne researchers collaborating with ...
Elana Simon was 10 years old when she started to experience severe pains in her abdomen. For two years, puzzled doctors put forward diagnoses including lactose intolerance, Crohn's disease and ...
The Kentucky Tick Surveillance Project is now accepting tick-testing submissions from Kentucky residents. This project, produced by the University of Kentucky Martin-Gatton College of Agriculture, Food and Environment, aims to improve knowledge about where ticks are found and the diseases they might carry.. The results help further public health research at the state level and alleviate ...
The economic impact of federally sponsored research at The University of Texas at Arlington was $38 million in 2022, with expenditures spread among 725 unique vendors, according to a new report. Of that total, the University spent about $24 million on research-related goods and services in Texas.