- MIT-IBM Watson AI Lab
- Inside the lab

Papers + Code
IBM Research
314 Main St. Cambridge, MA 02141
- NeurIPS 2020
- NeurIPS 2021
- What’s Next in AI
Peer-review is the lifeblood of scientific validation and a guardrail against runaway hype in AI. Our commitment to publishing in the top venues reflects our grounding in what is real, reproducible, and truly innovative.
Subscribe to the PwC Newsletter
Join the community, latest research, positivity violations in marginal structural survival models with time-dependent confounding: a simulation study on iptw-estimator performance.
mspreafico/posviolmsm • 28 Mar 2024
In longitudinal observational studies, marginal structural models (MSMs) are a class of causal models used to analyze the effect of an exposure on the (survival) outcome of interest while accounting for exposure-affected time-dependent confounding.
Methodology
Cumulative Incidence Function Estimation Based on Population-Based Biobank Data
david-zucker/illness-death • 27 Mar 2024
Many countries have established population-based biobanks, which are being used increasingly in epidemiolgical and clinical research.
Modifying Gibbs sampling to avoid self transitions
radfordneal/gibbsmod • 26 Mar 2024
Methods that do reduce the probability of self transitions to the minimum, but do not satisfy the conditions of Peskun's theorem, have also been devised, by Suwa and Todo.
Computation Computational Physics
Personalized Imputation in metric spaces via conformal prediction: Applications in Predicting Diabetes Development with Continuous Glucose Monitoring Information
carladiaz/conformal_imputation • 26 Mar 2024
The aim of this paper is to introduce a novel two-step framework that comprises: (i) a imputation methods for statistical objects taking values in metrics spaces, and (ii) a criterion for personalizing imputation using conformal inference techniques.
Methodology Applications
Assessing COVID-19 Vaccine Effectiveness in Observational Studies via Nested Trial Emulation
justindemonte/ve_nte • 26 Mar 2024
Typically, when nested trial emulation (NTE) is employed, effect estimates are pooled across trials to increase statistical efficiency.
Measuring Dependence between Events
jan-lukas-wermuth/bcor • 26 Mar 2024
Countless such dependence measures exist, but there is little theoretical guidance on how they compare and on their advantages and shortcomings.
Testing for sufficient follow-up in survival data with a cure fraction
tp-yuen/curesfutest • 25 Mar 2024
In order to estimate the proportion of `immune' or `cured' subjects who will never experience failure, a sufficiently long follow-up period is required.
Optimized Model Selection for Estimating Treatment Effects from Costly Simulations of the US Opioid Epidemic
abdulrahmanfci/model-selection • 23 Mar 2024
In this paper, we discuss different methods to explore what model works best at a specific sample size.
Methodology Multiagent Systems Social and Information Networks Applications
Computationally Scalable Bayesian SPDE Modeling for Censored Spatial Responses
SumanM47/CensSpBayes • 23 Mar 2024
Finally, the fit of this fully Bayesian model to the concentration of PFOS in groundwater available at 24, 959 sites across California, where 46. 62\% responses are censored, produces prediction surface and uncertainty quantification in real time, thereby substantiating the applicability and scalability of the proposed method.
Methodology Applications Computation
Augmented Doubly Robust Post-Imputation Inference for Proteomic Data
Quantitative measurements produced by mass spectrometry proteomics experiments offer a direct way to explore the role of proteins in molecular mechanisms.
Subscribe to the PwC Newsletter
Join the community, latest research, haina storage: a decentralized secure storage framework based on improved blockchain structure.
Zijian-Zhou/Haina_Storage_Exp • 2 Apr 2024
By introducing the network environment as an essential evaluation parameter of storage right decision, the energy and time consumption of decision-making are reduced, and the fairness of decision-making is improved.
Cryptography and Security Distributed, Parallel, and Cluster Computing
Design, Fabrication and Evaluation of a Stretchable High-Density Electromyography Array
rejinjohnvarghese/stretchable-hmi-array • 29 Mar 2024
The adoption of high-density electrode systems for human-machine interfaces in real-life applications has been impeded by practical and technical challenges, including noise interference, motion artifacts and the lack of compact electrode interfaces.
A Public and Reproducible Assessment of the Topics API on Real Data
yohhaan/topics_api_analysis • 28 Mar 2024
The central point of contention is largely around the realism of the datasets used in these analyses and their reproducibility; researchers using data collected on a small sample of users or generating synthetic datasets, while Google's results are inferred from a private dataset.
Cryptography and Security
Deep CSI Compression for Dual-Polarized Massive MIMO Channels with Disentangled Representation Learning

In practical dual-polarized MIMO scenarios, channels in the vertical and horizontal polarization directions tend to exhibit high polarization correlation.
Information Theory Signal Processing Information Theory
Cleaning data with Swipe
antoonbronselaer/swipe-reproducibility • 28 Mar 2024
In this paper, we explore an extreme variant of this idea in which the Chase tree has only one path.
Uncover the Premeditated Attacks: Detecting Exploitable Reentrancy Vulnerabilities by Identifying Attacker Contracts
shuo-young/blockwatchdog • 28 Mar 2024
Notably, only 18 of the identified 159 victim contracts can be reported by current reentrancy detection tools.
Cryptography and Security Software Engineering
Emotion Neural Transducer for Fine-Grained Speech Emotion Recognition
We first extend typical neural transducer with emotion joint network to construct emotion lattice for fine-grained SER.
Sound Audio and Speech Processing
Genos: General In-Network Unsupervised Intrusion Detection by Rule Extraction
ruoyu-li/genos-infocom24 • 28 Mar 2024
In this paper, we propose Genos, a general in-network framework for unsupervised A-NIDS by rule extraction, which consists of a Model Compiler, a Model Interpreter, and a Model Debugger.
UniFaaS: Programming across Distributed Cyberinfrastructure with Federated Function Serving
sustech-hpclab/unifaas • 28 Mar 2024
We show that UniFaaS can improve the performance of a real-world drug screening workflow by as much as 22. 99% when employing an additional 19. 48% of resources and a montage workflow by 54. 41% when employing an additional 47. 83% of resources across multiple distributed clusters, in contrast to using a single cluster
Distributed, Parallel, and Cluster Computing
Learning Sampling Distribution and Safety Filter for Autonomous Driving with VQ-VAE and Differentiable Optimization

We use backpropagation through the optimization layers in a self-supervised learning set-up to learn good initialization and optimal parameters of the safety filter.
Search code, repositories, users, issues, pull requests...
Provide feedback.
We read every piece of feedback, and take your input very seriously.
Saved searches
Use saved searches to filter your results more quickly.
To see all available qualifiers, see our documentation .
- Notifications
Tips for releasing research code in Machine Learning (with official NeurIPS 2020 recommendations)
paperswithcode/releasing-research-code
Folders and files, repository files navigation, tips for publishing research code.
💡 Collated best practices from most popular ML research repositories - now official guidelines at NeurIPS 2021!
Based on analysis of more than 200 Machine Learning repositories, these recommendations facilitate reproducibility and correlate with GitHub stars - for more details, see our our blog post .
For NeurIPS 2021 code submissions it is recommended (but not mandatory) to use the README.md template and check as many items on the ML Code Completeness Checklist (described below) as possible.
📋 README.md template
We provide a README.md template that you can use for releasing ML research repositories. The sections in the template were derived by looking at existing repositories, seeing which had the best reception in the community, and then looking at common components that correlate with popularity.
✓ ML Code Completeness Checklist
We compiled this checklist by looking at what's common to the most popular ML research repositories. In addition, we prioritized items that facilitate reproducibility and make it easier for others build upon research code.
The ML Code Completeness Checklist consists of five items:
- Specification of dependencies
- Training code
- Evaluation code
- Pre-trained models
- README file including table of results accompanied by precise commands to run/produce those results
We verified that repositories that check more items on the checklist also tend to have a higher number of GitHub stars. This was verified by analysing official NeurIPS 2019 repositories - more details in the blog post . We also provide the data and notebook to reproduce this analysis from the post.
NeurIPS 2019 repositories that had all five of these components had the highest number of GitHub stars (median of 196 and mean of 2,664 stars).
We explain each item on the checklist in detail blow.
1. Specification of dependencies
If you are using Python, this means providing a requirements.txt file (if using pip and virtualenv ), providing environment.yml file (if using anaconda), or a setup.py if your code is a library.
It is good practice to provide a section in your README.md that explains how to install these dependencies. Assume minimal background knowledge and be clear and comprehensive - if users cannot set up your dependencies they are likely to give up on the rest of your code as well.
If you wish to provide whole reproducible environments, you might want to consider using Docker and upload a Docker image of your environment into Dockerhub.
2. Training code
Your code should have a training script that can be used to obtain the principal results stated in the paper. This means you should include hyperparameters and any tricks that were used in the process of getting your results. To maximize usefulness, ideally this code should be written with extensibility in mind: what if your user wants to use the same training script on their own dataset?
You can provide a documented command line wrapper such as train.py to serve as a useful entry point for your users.
3. Evaluation code
Model evaluation and experiments often depend on subtle details that are not always possible to explain in the paper. This is why including the exact code you used to evaluate or run experiments is helpful to give a complete description of the procedure. In turn, this helps the user to trust, understand and build on your research.
You can provide a documented command line wrapper such as eval.py to serve as a useful entry point for your users.
4. Pre-trained models
Training a model from scratch can be time-consuming and expensive. One way to increase trust in your results is to provide a pre-trained model that the community can evaluate to obtain the end results. This means users can see the results are credible without having to train afresh.
Another common use case is fine-tuning for downstream task, where it's useful to release a pretrained model so others can build on it for application to their own datasets.
Lastly, some users might want to try out your model to see if it works on some example data. Providing pre-trained models allows your users to play around with your work and aids understanding of the paper's achievements.
5. README file includes table of results accompanied by precise command to run to produce those results
Adding a table of results into README.md lets your users quickly understand what to expect from the repository (see the README.md template for an example). Instructions on how to reproduce those results (with links to any relevant scripts, pretrained models etc) can provide another entry point for the user and directly facilitate reproducibility. In some cases, the main result of a paper is a Figure, but that might be more difficult for users to understand without reading the paper.
You can further help the user understand and contextualize your results by linking back to the full leaderboard that has up-to-date results from other papers. There are multiple leaderboard services where this information is stored.
🎉 Additional awesome resources for releasing research code
Hosting pretrained models files.
- Zenodo - versioning, 50GB, free bandwidth, DOI, provides long-term preservation
- GitHub Releases - versioning, 2GB file limit, free bandwidth
- OneDrive - versioning, 2GB (free)/ 1TB (with Office 365), free bandwidth
- Google Drive - versioning, 15GB, free bandwidth
- Dropbox - versioning, 2GB (paid unlimited), free bandwidth
- AWS S3 - versioning, paid only, paid bandwidth
- huggingface_hub - versioning, no size limitations, free bandwidth
- DAGsHub - versioning, no size limitations, free bandwith
- CodaLab Worksheets - 10GB, free bandwith
Managing model files
- RClone - provides unified access to many different cloud storage providers
Standardized model interfaces
- PyTorch Hub
- Tensorflow Hub
- Hugging Face NLP models
Results leaderboards
- Papers with Code leaderboards - with 4000+ leaderboards
- CodaLab Competitions - with 450+ leaderboards
- EvalAI - with 100+ leaderboards
- NLP Progress - with 90+ leaderboards
- Collective Knowledge - with 40+ leaderboards
- Weights & Biases - Benchmarks - with 9+ leaderboards
Making project pages
- GitHub pages
Making demos, tutorials, executable papers
- Google Colab
- CodaLab Worksheets
Contributing
If you'd like to contribute, or have any suggestions for these guidelines, you can contact us at [email protected] or open an issue on this GitHub repository.
All contributions welcome! All content in this repository is licensed under the MIT license.
Contributors 9
Instant access to code, for any arXiv paper
- Author By ame5
- Publication date December 10, 2020
- Categories: arXiv updates , arXivLabs
In October, arXiv released a new feature empowering arXiv authors to link their Machine Learning articles to associated code . Developed in an arXivLabs collaboration with Papers with Code , the tool was met with great enthusiasm from arXiv’s ML community.
Now, we’re expanding the capability beyond Machine Learning to arXiv papers in every category. And, to better align with our arXiv communities, PwC is launching new sites in computer science, physics, mathematics, astronomy and statistics to help researchers explore code in these fields.
As part of this expansion, PwC is indexing 600,000 additional papers, automatically detecting and linking to the code libraries for papers in categories beyond Machine Learning and also continuing to identify code, results, and methods within ML.
This expansion was driven by the broader research community’s demand. After the October release, researchers beyond ML expressed a desire for easy access to relevant code. Researchers in physics, astronomy, and other fields began adding their work to PwC. To answer the popular demand, we worked together with PwC to formally support additional fields.
arXiv authors and readers value speed and openness, and instant access to publicly available code supports both.
Subscribe By Email
Get every new post delivered right to your inbox.
Your Email Leave this field blank
This form is protected by reCAPTCHA and the Google Privacy Policy and Terms of Service apply.
Qualitative Data Coding 101
How to code qualitative data, the smart way (with examples).
By: Jenna Crosley (PhD) | Reviewed by:Dr Eunice Rautenbach | December 2020
As we’ve discussed previously , qualitative research makes use of non-numerical data – for example, words, phrases or even images and video. To analyse this kind of data, the first dragon you’ll need to slay is qualitative data coding (or just “coding” if you want to sound cool). But what exactly is coding and how do you do it?
Overview: Qualitative Data Coding
In this post, we’ll explain qualitative data coding in simple terms. Specifically, we’ll dig into:
- What exactly qualitative data coding is
- What different types of coding exist
- How to code qualitative data (the process)
- Moving from coding to qualitative analysis
- Tips and tricks for quality data coding
What is qualitative data coding?
Let’s start by understanding what a code is. At the simplest level, a code is a label that describes the content of a piece of text. For example, in the sentence:
“Pigeons attacked me and stole my sandwich.”
You could use “pigeons” as a code. This code simply describes that the sentence involves pigeons.
So, building onto this, qualitative data coding is the process of creating and assigning codes to categorise data extracts. You’ll then use these codes later down the road to derive themes and patterns for your qualitative analysis (for example, thematic analysis ). Coding and analysis can take place simultaneously, but it’s important to note that coding does not necessarily involve identifying themes (depending on which textbook you’re reading, of course). Instead, it generally refers to the process of labelling and grouping similar types of data to make generating themes and analysing the data more manageable.
Makes sense? Great. But why should you bother with coding at all? Why not just look for themes from the outset? Well, coding is a way of making sure your data is valid . In other words, it helps ensure that your analysis is undertaken systematically and that other researchers can review it (in the world of research, we call this transparency). In other words, good coding is the foundation of high-quality analysis.

What are the different types of coding?
Now that we’ve got a plain-language definition of coding on the table, the next step is to understand what types of coding exist. Let’s start with the two main approaches, deductive and inductive coding.
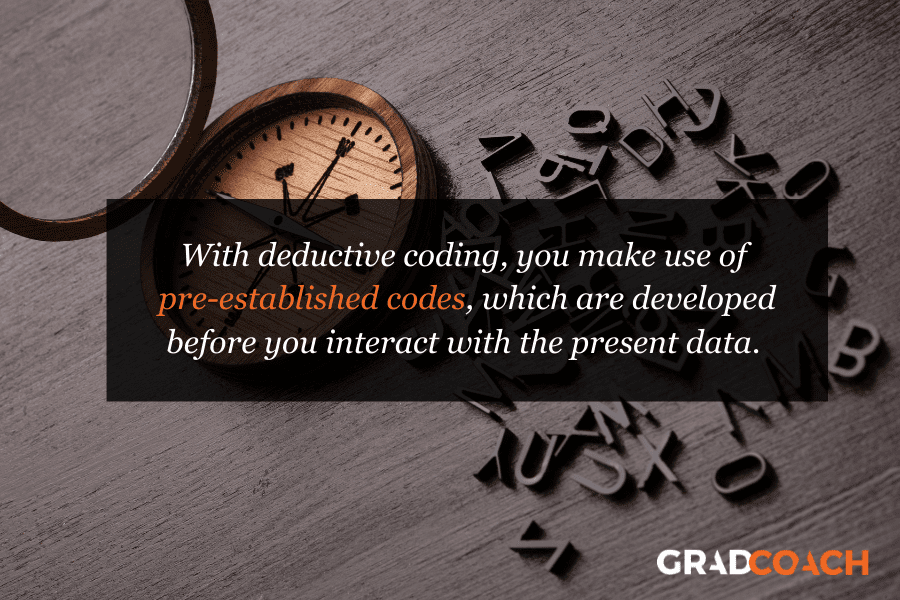
Deductive coding 101
With deductive coding, we make use of pre-established codes, which are developed before you interact with the present data. This usually involves drawing up a set of codes based on a research question or previous research . You could also use a code set from the codebook of a previous study.
For example, if you were studying the eating habits of college students, you might have a research question along the lines of
“What foods do college students eat the most?”
As a result of this research question, you might develop a code set that includes codes such as “sushi”, “pizza”, and “burgers”.
Deductive coding allows you to approach your analysis with a very tightly focused lens and quickly identify relevant data . Of course, the downside is that you could miss out on some very valuable insights as a result of this tight, predetermined focus.
Inductive coding 101
But what about inductive coding? As we touched on earlier, this type of coding involves jumping right into the data and then developing the codes based on what you find within the data.
For example, if you were to analyse a set of open-ended interviews , you wouldn’t necessarily know which direction the conversation would flow. If a conversation begins with a discussion of cats, it may go on to include other animals too, and so you’d add these codes as you progress with your analysis. Simply put, with inductive coding, you “go with the flow” of the data.
Inductive coding is great when you’re researching something that isn’t yet well understood because the coding derived from the data helps you explore the subject. Therefore, this type of coding is usually used when researchers want to investigate new ideas or concepts , or when they want to create new theories.
A little bit of both… hybrid coding approaches
If you’ve got a set of codes you’ve derived from a research topic, literature review or a previous study (i.e. a deductive approach), but you still don’t have a rich enough set to capture the depth of your qualitative data, you can combine deductive and inductive methods – this is called a hybrid coding approach.
To adopt a hybrid approach, you’ll begin your analysis with a set of a priori codes (deductive) and then add new codes (inductive) as you work your way through the data. Essentially, the hybrid coding approach provides the best of both worlds, which is why it’s pretty common to see this in research.
Need a helping hand?
How to code qualitative data
Now that we’ve looked at the main approaches to coding, the next question you’re probably asking is “how do I actually do it?”. Let’s take a look at the coding process , step by step.
Both inductive and deductive methods of coding typically occur in two stages: initial coding and line by line coding .
In the initial coding stage, the objective is to get a general overview of the data by reading through and understanding it. If you’re using an inductive approach, this is also where you’ll develop an initial set of codes. Then, in the second stage (line by line coding), you’ll delve deeper into the data and (re)organise it according to (potentially new) codes.
Step 1 – Initial coding
The first step of the coding process is to identify the essence of the text and code it accordingly. While there are various qualitative analysis software packages available, you can just as easily code textual data using Microsoft Word’s “comments” feature.
Let’s take a look at a practical example of coding. Assume you had the following interview data from two interviewees:
What pets do you have?
I have an alpaca and three dogs.
Only one alpaca? They can die of loneliness if they don’t have a friend.
I didn’t know that! I’ll just have to get five more.
I have twenty-three bunnies. I initially only had two, I’m not sure what happened.
In the initial stage of coding, you could assign the code of “pets” or “animals”. These are just initial, fairly broad codes that you can (and will) develop and refine later. In the initial stage, broad, rough codes are fine – they’re just a starting point which you will build onto in the second stage.
How to decide which codes to use
But how exactly do you decide what codes to use when there are many ways to read and interpret any given sentence? Well, there are a few different approaches you can adopt. The main approaches to initial coding include:
- In vivo coding
Process coding
- Open coding
Descriptive coding
Structural coding.
- Value coding
Let’s take a look at each of these:
In vivo coding
When you use in vivo coding, you make use of a participants’ own words , rather than your interpretation of the data. In other words, you use direct quotes from participants as your codes. By doing this, you’ll avoid trying to infer meaning, rather staying as close to the original phrases and words as possible.
In vivo coding is particularly useful when your data are derived from participants who speak different languages or come from different cultures. In these cases, it’s often difficult to accurately infer meaning due to linguistic or cultural differences.
For example, English speakers typically view the future as in front of them and the past as behind them. However, this isn’t the same in all cultures. Speakers of Aymara view the past as in front of them and the future as behind them. Why? Because the future is unknown, so it must be out of sight (or behind us). They know what happened in the past, so their perspective is that it’s positioned in front of them, where they can “see” it.
In a scenario like this one, it’s not possible to derive the reason for viewing the past as in front and the future as behind without knowing the Aymara culture’s perception of time. Therefore, in vivo coding is particularly useful, as it avoids interpretation errors.
Next up, there’s process coding, which makes use of action-based codes . Action-based codes are codes that indicate a movement or procedure. These actions are often indicated by gerunds (words ending in “-ing”) – for example, running, jumping or singing.
Process coding is useful as it allows you to code parts of data that aren’t necessarily spoken, but that are still imperative to understanding the meaning of the texts.
An example here would be if a participant were to say something like, “I have no idea where she is”. A sentence like this can be interpreted in many different ways depending on the context and movements of the participant. The participant could shrug their shoulders, which would indicate that they genuinely don’t know where the girl is; however, they could also wink, showing that they do actually know where the girl is.
Simply put, process coding is useful as it allows you to, in a concise manner, identify the main occurrences in a set of data and provide a dynamic account of events. For example, you may have action codes such as, “describing a panda”, “singing a song about bananas”, or “arguing with a relative”.

Descriptive coding aims to summarise extracts by using a single word or noun that encapsulates the general idea of the data. These words will typically describe the data in a highly condensed manner, which allows the researcher to quickly refer to the content.
Descriptive coding is very useful when dealing with data that appear in forms other than traditional text – i.e. video clips, sound recordings or images. For example, a descriptive code could be “food” when coding a video clip that involves a group of people discussing what they ate throughout the day, or “cooking” when coding an image showing the steps of a recipe.
Structural coding involves labelling and describing specific structural attributes of the data. Generally, it includes coding according to answers to the questions of “ who ”, “ what ”, “ where ”, and “ how ”, rather than the actual topics expressed in the data. This type of coding is useful when you want to access segments of data quickly, and it can help tremendously when you’re dealing with large data sets.
For example, if you were coding a collection of theses or dissertations (which would be quite a large data set), structural coding could be useful as you could code according to different sections within each of these documents – i.e. according to the standard dissertation structure . What-centric labels such as “hypothesis”, “literature review”, and “methodology” would help you to efficiently refer to sections and navigate without having to work through sections of data all over again.
Structural coding is also useful for data from open-ended surveys. This data may initially be difficult to code as they lack the set structure of other forms of data (such as an interview with a strict set of questions to be answered). In this case, it would useful to code sections of data that answer certain questions such as “who?”, “what?”, “where?” and “how?”.
Let’s take a look at a practical example. If we were to send out a survey asking people about their dogs, we may end up with a (highly condensed) response such as the following:
Bella is my best friend. When I’m at home I like to sit on the floor with her and roll her ball across the carpet for her to fetch and bring back to me. I love my dog.
In this set, we could code Bella as “who”, dog as “what”, home and floor as “where”, and roll her ball as “how”.
Values coding
Finally, values coding involves coding that relates to the participant’s worldviews . Typically, this type of coding focuses on excerpts that reflect the values, attitudes, and beliefs of the participants. Values coding is therefore very useful for research exploring cultural values and intrapersonal and experiences and actions.
To recap, the aim of initial coding is to understand and familiarise yourself with your data , to develop an initial code set (if you’re taking an inductive approach) and to take the first shot at coding your data . The coding approaches above allow you to arrange your data so that it’s easier to navigate during the next stage, line by line coding (we’ll get to this soon).
While these approaches can all be used individually, it’s important to remember that it’s possible, and potentially beneficial, to combine them . For example, when conducting initial coding with interviews, you could begin by using structural coding to indicate who speaks when. Then, as a next step, you could apply descriptive coding so that you can navigate to, and between, conversation topics easily.
Step 2 – Line by line coding
Once you’ve got an overall idea of our data, are comfortable navigating it and have applied some initial codes, you can move on to line by line coding. Line by line coding is pretty much exactly what it sounds like – reviewing your data, line by line, digging deeper and assigning additional codes to each line.
With line-by-line coding, the objective is to pay close attention to your data to add detail to your codes. For example, if you have a discussion of beverages and you previously just coded this as “beverages”, you could now go deeper and code more specifically, such as “coffee”, “tea”, and “orange juice”. The aim here is to scratch below the surface. This is the time to get detailed and specific so as to capture as much richness from the data as possible.
In the line-by-line coding process, it’s useful to code everything in your data, even if you don’t think you’re going to use it (you may just end up needing it!). As you go through this process, your coding will become more thorough and detailed, and you’ll have a much better understanding of your data as a result of this, which will be incredibly valuable in the analysis phase.


Moving from coding to analysis
Once you’ve completed your initial coding and line by line coding, the next step is to start your analysis . Of course, the coding process itself will get you in “analysis mode” and you’ll probably already have some insights and ideas as a result of it, so you should always keep notes of your thoughts as you work through the coding.
When it comes to qualitative data analysis, there are many different types of analyses (we discuss some of the most popular ones here ) and the type of analysis you adopt will depend heavily on your research aims, objectives and questions . Therefore, we’re not going to go down that rabbit hole here, but we’ll cover the important first steps that build the bridge from qualitative data coding to qualitative analysis.
When starting to think about your analysis, it’s useful to ask yourself the following questions to get the wheels turning:
- What actions are shown in the data?
- What are the aims of these interactions and excerpts? What are the participants potentially trying to achieve?
- How do participants interpret what is happening, and how do they speak about it? What does their language reveal?
- What are the assumptions made by the participants?
- What are the participants doing? What is going on?
- Why do I want to learn about this? What am I trying to find out?
- Why did I include this particular excerpt? What does it represent and how?

Code categorisation
Categorisation is simply the process of reviewing everything you’ve coded and then creating code categories that can be used to guide your future analysis. In other words, it’s about creating categories for your code set. Let’s take a look at a practical example.
If you were discussing different types of animals, your initial codes may be “dogs”, “llamas”, and “lions”. In the process of categorisation, you could label (categorise) these three animals as “mammals”, whereas you could categorise “flies”, “crickets”, and “beetles” as “insects”. By creating these code categories, you will be making your data more organised, as well as enriching it so that you can see new connections between different groups of codes.
Theme identification
From the coding and categorisation processes, you’ll naturally start noticing themes. Therefore, the logical next step is to identify and clearly articulate the themes in your data set. When you determine themes, you’ll take what you’ve learned from the coding and categorisation and group it all together to develop themes. This is the part of the coding process where you’ll try to draw meaning from your data, and start to produce a narrative . The nature of this narrative depends on your research aims and objectives, as well as your research questions (sounds familiar?) and the qualitative data analysis method you’ve chosen, so keep these factors front of mind as you scan for themes.

Tips & tricks for quality coding
Before we wrap up, let’s quickly look at some general advice, tips and suggestions to ensure your qualitative data coding is top-notch.
- Before you begin coding, plan out the steps you will take and the coding approach and technique(s) you will follow to avoid inconsistencies.
- When adopting deductive coding, it’s useful to use a codebook from the start of the coding process. This will keep your work organised and will ensure that you don’t forget any of your codes.
- Whether you’re adopting an inductive or deductive approach, keep track of the meanings of your codes and remember to revisit these as you go along.
- Avoid using synonyms for codes that are similar, if not the same. This will allow you to have a more uniform and accurate coded dataset and will also help you to not get overwhelmed by your data.
- While coding, make sure that you remind yourself of your aims and coding method. This will help you to avoid directional drift , which happens when coding is not kept consistent.
- If you are working in a team, make sure that everyone has been trained and understands how codes need to be assigned.

Psst… there’s more (for free)
This post is part of our dissertation mini-course, which covers everything you need to get started with your dissertation, thesis or research project.
You Might Also Like:

30 Comments
I appreciated the valuable information provided to accomplish the various stages of the inductive and inductive coding process. However, I would have been extremely satisfied to be appraised of the SPECIFIC STEPS to follow for: 1. Deductive coding related to the phenomenon and its features to generate the codes, categories, and themes. 2. Inductive coding related to using (a) Initial (b) Axial, and (c) Thematic procedures using transcribe data from the research questions
Thank you so much for this. Very clear and simplified discussion about qualitative data coding.
This is what I want and the way I wanted it. Thank you very much.
All of the information’s are valuable and helpful. Thank for you giving helpful information’s. Can do some article about alternative methods for continue researches during the pandemics. It is more beneficial for those struggling to continue their researchers.
Thank you for your information on coding qualitative data, this is a very important point to be known, really thank you very much.
Very useful article. Clear, articulate and easy to understand. Thanks
This is very useful. You have simplified it the way I wanted it to be! Thanks
Thank you so very much for explaining, this is quite helpful!
hello, great article! well written and easy to understand. Can you provide some of the sources in this article used for further reading purposes?
You guys are doing a great job out there . I will not realize how many students you help through your articles and post on a daily basis. I have benefited a lot from your work. this is remarkable.
Wonderful one thank you so much.
Hello, I am doing qualitative research, please assist with example of coding format.
This is an invaluable website! Thank you so very much!
Well explained and easy to follow the presentation. A big thumbs up to you. Greatly appreciate the effort 👏👏👏👏
Thank you for this clear article with examples
Thank you for the detailed explanation. I appreciate your great effort. Congrats!
Ahhhhhhhhhh! You just killed me with your explanation. Crystal clear. Two Cheers!
D0 you have primary references that was used when creating this? If so, can you share them?
Being a complete novice to the field of qualitative data analysis, your indepth analysis of the process of thematic analysis has given me better insight. Thank you so much.
Excellent summary
Thank you so much for your precise and very helpful information about coding in qualitative data.
Thanks a lot to this helpful information. You cleared the fog in my brain.
Glad to hear that!
This has been very helpful. I am excited and grateful.
I still don’t understand the coding and categorizing of qualitative research, please give an example on my research base on the state of government education infrastructure environment in PNG
Wahho, this is amazing and very educational to have come across this site.. from a little search to a wide discovery of knowledge.
Thanks I really appreciate this.
Thank you so much! Very grateful.
This was truly helpful. I have been so lost, and this simplified the process for me.
Just at the right time when I needed to distinguish between inductive and
deductive data analysis of my Focus group discussion results very helpful
Submit a Comment Cancel reply
Your email address will not be published. Required fields are marked *
Save my name, email, and website in this browser for the next time I comment.
- Print Friendly
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 26 March 2024
Predicting and improving complex beer flavor through machine learning
- Michiel Schreurs ORCID: orcid.org/0000-0002-9449-5619 1 , 2 , 3 na1 ,
- Supinya Piampongsant 1 , 2 , 3 na1 ,
- Miguel Roncoroni ORCID: orcid.org/0000-0001-7461-1427 1 , 2 , 3 na1 ,
- Lloyd Cool ORCID: orcid.org/0000-0001-9936-3124 1 , 2 , 3 , 4 ,
- Beatriz Herrera-Malaver ORCID: orcid.org/0000-0002-5096-9974 1 , 2 , 3 ,
- Christophe Vanderaa ORCID: orcid.org/0000-0001-7443-5427 4 ,
- Florian A. Theßeling 1 , 2 , 3 ,
- Łukasz Kreft ORCID: orcid.org/0000-0001-7620-4657 5 ,
- Alexander Botzki ORCID: orcid.org/0000-0001-6691-4233 5 ,
- Philippe Malcorps 6 ,
- Luk Daenen 6 ,
- Tom Wenseleers ORCID: orcid.org/0000-0002-1434-861X 4 &
- Kevin J. Verstrepen ORCID: orcid.org/0000-0002-3077-6219 1 , 2 , 3
Nature Communications volume 15 , Article number: 2368 ( 2024 ) Cite this article
44k Accesses
797 Altmetric
Metrics details
- Chemical engineering
- Gas chromatography
- Machine learning
- Metabolomics
- Taste receptors
The perception and appreciation of food flavor depends on many interacting chemical compounds and external factors, and therefore proves challenging to understand and predict. Here, we combine extensive chemical and sensory analyses of 250 different beers to train machine learning models that allow predicting flavor and consumer appreciation. For each beer, we measure over 200 chemical properties, perform quantitative descriptive sensory analysis with a trained tasting panel and map data from over 180,000 consumer reviews to train 10 different machine learning models. The best-performing algorithm, Gradient Boosting, yields models that significantly outperform predictions based on conventional statistics and accurately predict complex food features and consumer appreciation from chemical profiles. Model dissection allows identifying specific and unexpected compounds as drivers of beer flavor and appreciation. Adding these compounds results in variants of commercial alcoholic and non-alcoholic beers with improved consumer appreciation. Together, our study reveals how big data and machine learning uncover complex links between food chemistry, flavor and consumer perception, and lays the foundation to develop novel, tailored foods with superior flavors.
Similar content being viewed by others
BitterSweet: Building machine learning models for predicting the bitter and sweet taste of small molecules
Rudraksh Tuwani, Somin Wadhwa & Ganesh Bagler
Sensory lexicon and aroma volatiles analysis of brewing malt
Xiaoxia Su, Miao Yu, … Tianyi Du
Predicting odor from molecular structure: a multi-label classification approach
Kushagra Saini & Venkatnarayan Ramanathan
Introduction
Predicting and understanding food perception and appreciation is one of the major challenges in food science. Accurate modeling of food flavor and appreciation could yield important opportunities for both producers and consumers, including quality control, product fingerprinting, counterfeit detection, spoilage detection, and the development of new products and product combinations (food pairing) 1 , 2 , 3 , 4 , 5 , 6 . Accurate models for flavor and consumer appreciation would contribute greatly to our scientific understanding of how humans perceive and appreciate flavor. Moreover, accurate predictive models would also facilitate and standardize existing food assessment methods and could supplement or replace assessments by trained and consumer tasting panels, which are variable, expensive and time-consuming 7 , 8 , 9 . Lastly, apart from providing objective, quantitative, accurate and contextual information that can help producers, models can also guide consumers in understanding their personal preferences 10 .
Despite the myriad of applications, predicting food flavor and appreciation from its chemical properties remains a largely elusive goal in sensory science, especially for complex food and beverages 11 , 12 . A key obstacle is the immense number of flavor-active chemicals underlying food flavor. Flavor compounds can vary widely in chemical structure and concentration, making them technically challenging and labor-intensive to quantify, even in the face of innovations in metabolomics, such as non-targeted metabolic fingerprinting 13 , 14 . Moreover, sensory analysis is perhaps even more complicated. Flavor perception is highly complex, resulting from hundreds of different molecules interacting at the physiochemical and sensorial level. Sensory perception is often non-linear, characterized by complex and concentration-dependent synergistic and antagonistic effects 15 , 16 , 17 , 18 , 19 , 20 , 21 that are further convoluted by the genetics, environment, culture and psychology of consumers 22 , 23 , 24 . Perceived flavor is therefore difficult to measure, with problems of sensitivity, accuracy, and reproducibility that can only be resolved by gathering sufficiently large datasets 25 . Trained tasting panels are considered the prime source of quality sensory data, but require meticulous training, are low throughput and high cost. Public databases containing consumer reviews of food products could provide a valuable alternative, especially for studying appreciation scores, which do not require formal training 25 . Public databases offer the advantage of amassing large amounts of data, increasing the statistical power to identify potential drivers of appreciation. However, public datasets suffer from biases, including a bias in the volunteers that contribute to the database, as well as confounding factors such as price, cult status and psychological conformity towards previous ratings of the product.
Classical multivariate statistics and machine learning methods have been used to predict flavor of specific compounds by, for example, linking structural properties of a compound to its potential biological activities or linking concentrations of specific compounds to sensory profiles 1 , 26 . Importantly, most previous studies focused on predicting organoleptic properties of single compounds (often based on their chemical structure) 27 , 28 , 29 , 30 , 31 , 32 , 33 , thus ignoring the fact that these compounds are present in a complex matrix in food or beverages and excluding complex interactions between compounds. Moreover, the classical statistics commonly used in sensory science 34 , 35 , 36 , 37 , 38 , 39 require a large sample size and sufficient variance amongst predictors to create accurate models. They are not fit for studying an extensive set of hundreds of interacting flavor compounds, since they are sensitive to outliers, have a high tendency to overfit and are less suited for non-linear and discontinuous relationships 40 .
In this study, we combine extensive chemical analyses and sensory data of a set of different commercial beers with machine learning approaches to develop models that predict taste, smell, mouthfeel and appreciation from compound concentrations. Beer is particularly suited to model the relationship between chemistry, flavor and appreciation. First, beer is a complex product, consisting of thousands of flavor compounds that partake in complex sensory interactions 41 , 42 , 43 . This chemical diversity arises from the raw materials (malt, yeast, hops, water and spices) and biochemical conversions during the brewing process (kilning, mashing, boiling, fermentation, maturation and aging) 44 , 45 . Second, the advent of the internet saw beer consumers embrace online review platforms, such as RateBeer (ZX Ventures, Anheuser-Busch InBev SA/NV) and BeerAdvocate (Next Glass, inc.). In this way, the beer community provides massive data sets of beer flavor and appreciation scores, creating extraordinarily large sensory databases to complement the analyses of our professional sensory panel. Specifically, we characterize over 200 chemical properties of 250 commercial beers, spread across 22 beer styles, and link these to the descriptive sensory profiling data of a 16-person in-house trained tasting panel and data acquired from over 180,000 public consumer reviews. These unique and extensive datasets enable us to train a suite of machine learning models to predict flavor and appreciation from a beer’s chemical profile. Dissection of the best-performing models allows us to pinpoint specific compounds as potential drivers of beer flavor and appreciation. Follow-up experiments confirm the importance of these compounds and ultimately allow us to significantly improve the flavor and appreciation of selected commercial beers. Together, our study represents a significant step towards understanding complex flavors and reinforces the value of machine learning to develop and refine complex foods. In this way, it represents a stepping stone for further computer-aided food engineering applications 46 .
To generate a comprehensive dataset on beer flavor, we selected 250 commercial Belgian beers across 22 different beer styles (Supplementary Fig. S1 ). Beers with ≤ 4.2% alcohol by volume (ABV) were classified as non-alcoholic and low-alcoholic. Blonds and Tripels constitute a significant portion of the dataset (12.4% and 11.2%, respectively) reflecting their presence on the Belgian beer market and the heterogeneity of beers within these styles. By contrast, lager beers are less diverse and dominated by a handful of brands. Rare styles such as Brut or Faro make up only a small fraction of the dataset (2% and 1%, respectively) because fewer of these beers are produced and because they are dominated by distinct characteristics in terms of flavor and chemical composition.
Extensive analysis identifies relationships between chemical compounds in beer
For each beer, we measured 226 different chemical properties, including common brewing parameters such as alcohol content, iso-alpha acids, pH, sugar concentration 47 , and over 200 flavor compounds (Methods, Supplementary Table S1 ). A large portion (37.2%) are terpenoids arising from hopping, responsible for herbal and fruity flavors 16 , 48 . A second major category are yeast metabolites, such as esters and alcohols, that result in fruity and solvent notes 48 , 49 , 50 . Other measured compounds are primarily derived from malt, or other microbes such as non- Saccharomyces yeasts and bacteria (‘wild flora’). Compounds that arise from spices or staling are labeled under ‘Others’. Five attributes (caloric value, total acids and total ester, hop aroma and sulfur compounds) are calculated from multiple individually measured compounds.
As a first step in identifying relationships between chemical properties, we determined correlations between the concentrations of the compounds (Fig. 1 , upper panel, Supplementary Data 1 and 2 , and Supplementary Fig. S2 . For the sake of clarity, only a subset of the measured compounds is shown in Fig. 1 ). Compounds of the same origin typically show a positive correlation, while absence of correlation hints at parameters varying independently. For example, the hop aroma compounds citronellol, and alpha-terpineol show moderate correlations with each other (Spearman’s rho=0.39 and 0.57), but not with the bittering hop component iso-alpha acids (Spearman’s rho=0.16 and −0.07). This illustrates how brewers can independently modify hop aroma and bitterness by selecting hop varieties and dosage time. If hops are added early in the boiling phase, chemical conversions increase bitterness while aromas evaporate, conversely, late addition of hops preserves aroma but limits bitterness 51 . Similarly, hop-derived iso-alpha acids show a strong anti-correlation with lactic acid and acetic acid, likely reflecting growth inhibition of lactic acid and acetic acid bacteria, or the consequent use of fewer hops in sour beer styles, such as West Flanders ales and Fruit beers, that rely on these bacteria for their distinct flavors 52 . Finally, yeast-derived esters (ethyl acetate, ethyl decanoate, ethyl hexanoate, ethyl octanoate) and alcohols (ethanol, isoamyl alcohol, isobutanol, and glycerol), correlate with Spearman coefficients above 0.5, suggesting that these secondary metabolites are correlated with the yeast genetic background and/or fermentation parameters and may be difficult to influence individually, although the choice of yeast strain may offer some control 53 .

Spearman rank correlations are shown. Descriptors are grouped according to their origin (malt (blue), hops (green), yeast (red), wild flora (yellow), Others (black)), and sensory aspect (aroma, taste, palate, and overall appreciation). Please note that for the chemical compounds, for the sake of clarity, only a subset of the total number of measured compounds is shown, with an emphasis on the key compounds for each source. For more details, see the main text and Methods section. Chemical data can be found in Supplementary Data 1 , correlations between all chemical compounds are depicted in Supplementary Fig. S2 and correlation values can be found in Supplementary Data 2 . See Supplementary Data 4 for sensory panel assessments and Supplementary Data 5 for correlation values between all sensory descriptors.
Interestingly, different beer styles show distinct patterns for some flavor compounds (Supplementary Fig. S3 ). These observations agree with expectations for key beer styles, and serve as a control for our measurements. For instance, Stouts generally show high values for color (darker), while hoppy beers contain elevated levels of iso-alpha acids, compounds associated with bitter hop taste. Acetic and lactic acid are not prevalent in most beers, with notable exceptions such as Kriek, Lambic, Faro, West Flanders ales and Flanders Old Brown, which use acid-producing bacteria ( Lactobacillus and Pediococcus ) or unconventional yeast ( Brettanomyces ) 54 , 55 . Glycerol, ethanol and esters show similar distributions across all beer styles, reflecting their common origin as products of yeast metabolism during fermentation 45 , 53 . Finally, low/no-alcohol beers contain low concentrations of glycerol and esters. This is in line with the production process for most of the low/no-alcohol beers in our dataset, which are produced through limiting fermentation or by stripping away alcohol via evaporation or dialysis, with both methods having the unintended side-effect of reducing the amount of flavor compounds in the final beer 56 , 57 .
Besides expected associations, our data also reveals less trivial associations between beer styles and specific parameters. For example, geraniol and citronellol, two monoterpenoids responsible for citrus, floral and rose flavors and characteristic of Citra hops, are found in relatively high amounts in Christmas, Saison, and Brett/co-fermented beers, where they may originate from terpenoid-rich spices such as coriander seeds instead of hops 58 .
Tasting panel assessments reveal sensorial relationships in beer
To assess the sensory profile of each beer, a trained tasting panel evaluated each of the 250 beers for 50 sensory attributes, including different hop, malt and yeast flavors, off-flavors and spices. Panelists used a tasting sheet (Supplementary Data 3 ) to score the different attributes. Panel consistency was evaluated by repeating 12 samples across different sessions and performing ANOVA. In 95% of cases no significant difference was found across sessions ( p > 0.05), indicating good panel consistency (Supplementary Table S2 ).
Aroma and taste perception reported by the trained panel are often linked (Fig. 1 , bottom left panel and Supplementary Data 4 and 5 ), with high correlations between hops aroma and taste (Spearman’s rho=0.83). Bitter taste was found to correlate with hop aroma and taste in general (Spearman’s rho=0.80 and 0.69), and particularly with “grassy” noble hops (Spearman’s rho=0.75). Barnyard flavor, most often associated with sour beers, is identified together with stale hops (Spearman’s rho=0.97) that are used in these beers. Lactic and acetic acid, which often co-occur, are correlated (Spearman’s rho=0.66). Interestingly, sweetness and bitterness are anti-correlated (Spearman’s rho = −0.48), confirming the hypothesis that they mask each other 59 , 60 . Beer body is highly correlated with alcohol (Spearman’s rho = 0.79), and overall appreciation is found to correlate with multiple aspects that describe beer mouthfeel (alcohol, carbonation; Spearman’s rho= 0.32, 0.39), as well as with hop and ester aroma intensity (Spearman’s rho=0.39 and 0.35).
Similar to the chemical analyses, sensorial analyses confirmed typical features of specific beer styles (Supplementary Fig. S4 ). For example, sour beers (Faro, Flanders Old Brown, Fruit beer, Kriek, Lambic, West Flanders ale) were rated acidic, with flavors of both acetic and lactic acid. Hoppy beers were found to be bitter and showed hop-associated aromas like citrus and tropical fruit. Malt taste is most detected among scotch, stout/porters, and strong ales, while low/no-alcohol beers, which often have a reputation for being ‘worty’ (reminiscent of unfermented, sweet malt extract) appear in the middle. Unsurprisingly, hop aromas are most strongly detected among hoppy beers. Like its chemical counterpart (Supplementary Fig. S3 ), acidity shows a right-skewed distribution, with the most acidic beers being Krieks, Lambics, and West Flanders ales.
Tasting panel assessments of specific flavors correlate with chemical composition
We find that the concentrations of several chemical compounds strongly correlate with specific aroma or taste, as evaluated by the tasting panel (Fig. 2 , Supplementary Fig. S5 , Supplementary Data 6 ). In some cases, these correlations confirm expectations and serve as a useful control for data quality. For example, iso-alpha acids, the bittering compounds in hops, strongly correlate with bitterness (Spearman’s rho=0.68), while ethanol and glycerol correlate with tasters’ perceptions of alcohol and body, the mouthfeel sensation of fullness (Spearman’s rho=0.82/0.62 and 0.72/0.57 respectively) and darker color from roasted malts is a good indication of malt perception (Spearman’s rho=0.54).

Heatmap colors indicate Spearman’s Rho. Axes are organized according to sensory categories (aroma, taste, mouthfeel, overall), chemical categories and chemical sources in beer (malt (blue), hops (green), yeast (red), wild flora (yellow), Others (black)). See Supplementary Data 6 for all correlation values.
Interestingly, for some relationships between chemical compounds and perceived flavor, correlations are weaker than expected. For example, the rose-smelling phenethyl acetate only weakly correlates with floral aroma. This hints at more complex relationships and interactions between compounds and suggests a need for a more complex model than simple correlations. Lastly, we uncovered unexpected correlations. For instance, the esters ethyl decanoate and ethyl octanoate appear to correlate slightly with hop perception and bitterness, possibly due to their fruity flavor. Iron is anti-correlated with hop aromas and bitterness, most likely because it is also anti-correlated with iso-alpha acids. This could be a sign of metal chelation of hop acids 61 , given that our analyses measure unbound hop acids and total iron content, or could result from the higher iron content in dark and Fruit beers, which typically have less hoppy and bitter flavors 62 .
Public consumer reviews complement expert panel data
To complement and expand the sensory data of our trained tasting panel, we collected 180,000 reviews of our 250 beers from the online consumer review platform RateBeer. This provided numerical scores for beer appearance, aroma, taste, palate, overall quality as well as the average overall score.
Public datasets are known to suffer from biases, such as price, cult status and psychological conformity towards previous ratings of a product. For example, prices correlate with appreciation scores for these online consumer reviews (rho=0.49, Supplementary Fig. S6 ), but not for our trained tasting panel (rho=0.19). This suggests that prices affect consumer appreciation, which has been reported in wine 63 , while blind tastings are unaffected. Moreover, we observe that some beer styles, like lagers and non-alcoholic beers, generally receive lower scores, reflecting that online reviewers are mostly beer aficionados with a preference for specialty beers over lager beers. In general, we find a modest correlation between our trained panel’s overall appreciation score and the online consumer appreciation scores (Fig. 3 , rho=0.29). Apart from the aforementioned biases in the online datasets, serving temperature, sample freshness and surroundings, which are all tightly controlled during the tasting panel sessions, can vary tremendously across online consumers and can further contribute to (among others, appreciation) differences between the two categories of tasters. Importantly, in contrast to the overall appreciation scores, for many sensory aspects the results from the professional panel correlated well with results obtained from RateBeer reviews. Correlations were highest for features that are relatively easy to recognize even for untrained tasters, like bitterness, sweetness, alcohol and malt aroma (Fig. 3 and below).
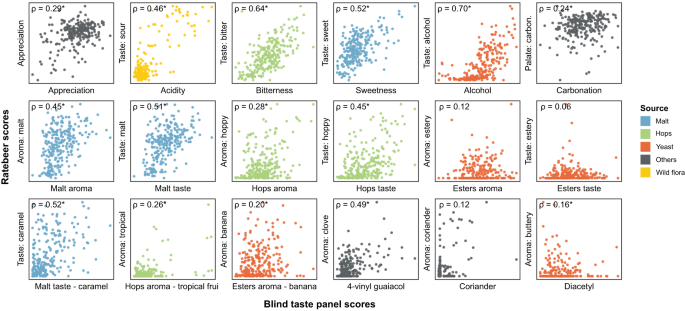
RateBeer text mining results can be found in Supplementary Data 7 . Rho values shown are Spearman correlation values, with asterisks indicating significant correlations ( p < 0.05, two-sided). All p values were smaller than 0.001, except for Esters aroma (0.0553), Esters taste (0.3275), Esters aroma—banana (0.0019), Coriander (0.0508) and Diacetyl (0.0134).
Besides collecting consumer appreciation from these online reviews, we developed automated text analysis tools to gather additional data from review texts (Supplementary Data 7 ). Processing review texts on the RateBeer database yielded comparable results to the scores given by the trained panel for many common sensory aspects, including acidity, bitterness, sweetness, alcohol, malt, and hop tastes (Fig. 3 ). This is in line with what would be expected, since these attributes require less training for accurate assessment and are less influenced by environmental factors such as temperature, serving glass and odors in the environment. Consumer reviews also correlate well with our trained panel for 4-vinyl guaiacol, a compound associated with a very characteristic aroma. By contrast, correlations for more specific aromas like ester, coriander or diacetyl are underrepresented in the online reviews, underscoring the importance of using a trained tasting panel and standardized tasting sheets with explicit factors to be scored for evaluating specific aspects of a beer. Taken together, our results suggest that public reviews are trustworthy for some, but not all, flavor features and can complement or substitute taste panel data for these sensory aspects.
Models can predict beer sensory profiles from chemical data
The rich datasets of chemical analyses, tasting panel assessments and public reviews gathered in the first part of this study provided us with a unique opportunity to develop predictive models that link chemical data to sensorial features. Given the complexity of beer flavor, basic statistical tools such as correlations or linear regression may not always be the most suitable for making accurate predictions. Instead, we applied different machine learning models that can model both simple linear and complex interactive relationships. Specifically, we constructed a set of regression models to predict (a) trained panel scores for beer flavor and quality and (b) public reviews’ appreciation scores from beer chemical profiles. We trained and tested 10 different models (Methods), 3 linear regression-based models (simple linear regression with first-order interactions (LR), lasso regression with first-order interactions (Lasso), partial least squares regressor (PLSR)), 5 decision tree models (AdaBoost regressor (ABR), extra trees (ET), gradient boosting regressor (GBR), random forest (RF) and XGBoost regressor (XGBR)), 1 support vector regression (SVR), and 1 artificial neural network (ANN) model.
To compare the performance of our machine learning models, the dataset was randomly split into a training and test set, stratified by beer style. After a model was trained on data in the training set, its performance was evaluated on its ability to predict the test dataset obtained from multi-output models (based on the coefficient of determination, see Methods). Additionally, individual-attribute models were ranked per descriptor and the average rank was calculated, as proposed by Korneva et al. 64 . Importantly, both ways of evaluating the models’ performance agreed in general. Performance of the different models varied (Table 1 ). It should be noted that all models perform better at predicting RateBeer results than results from our trained tasting panel. One reason could be that sensory data is inherently variable, and this variability is averaged out with the large number of public reviews from RateBeer. Additionally, all tree-based models perform better at predicting taste than aroma. Linear models (LR) performed particularly poorly, with negative R 2 values, due to severe overfitting (training set R 2 = 1). Overfitting is a common issue in linear models with many parameters and limited samples, especially with interaction terms further amplifying the number of parameters. L1 regularization (Lasso) successfully overcomes this overfitting, out-competing multiple tree-based models on the RateBeer dataset. Similarly, the dimensionality reduction of PLSR avoids overfitting and improves performance, to some extent. Still, tree-based models (ABR, ET, GBR, RF and XGBR) show the best performance, out-competing the linear models (LR, Lasso, PLSR) commonly used in sensory science 65 .
GBR models showed the best overall performance in predicting sensory responses from chemical information, with R 2 values up to 0.75 depending on the predicted sensory feature (Supplementary Table S4 ). The GBR models predict consumer appreciation (RateBeer) better than our trained panel’s appreciation (R 2 value of 0.67 compared to R 2 value of 0.09) (Supplementary Table S3 and Supplementary Table S4 ). ANN models showed intermediate performance, likely because neural networks typically perform best with larger datasets 66 . The SVR shows intermediate performance, mostly due to the weak predictions of specific attributes that lower the overall performance (Supplementary Table S4 ).
Model dissection identifies specific, unexpected compounds as drivers of consumer appreciation
Next, we leveraged our models to infer important contributors to sensory perception and consumer appreciation. Consumer preference is a crucial sensory aspects, because a product that shows low consumer appreciation scores often does not succeed commercially 25 . Additionally, the requirement for a large number of representative evaluators makes consumer trials one of the more costly and time-consuming aspects of product development. Hence, a model for predicting chemical drivers of overall appreciation would be a welcome addition to the available toolbox for food development and optimization.
Since GBR models on our RateBeer dataset showed the best overall performance, we focused on these models. Specifically, we used two approaches to identify important contributors. First, rankings of the most important predictors for each sensorial trait in the GBR models were obtained based on impurity-based feature importance (mean decrease in impurity). High-ranked parameters were hypothesized to be either the true causal chemical properties underlying the trait, to correlate with the actual causal properties, or to take part in sensory interactions affecting the trait 67 (Fig. 4A ). In a second approach, we used SHAP 68 to determine which parameters contributed most to the model for making predictions of consumer appreciation (Fig. 4B ). SHAP calculates parameter contributions to model predictions on a per-sample basis, which can be aggregated into an importance score.
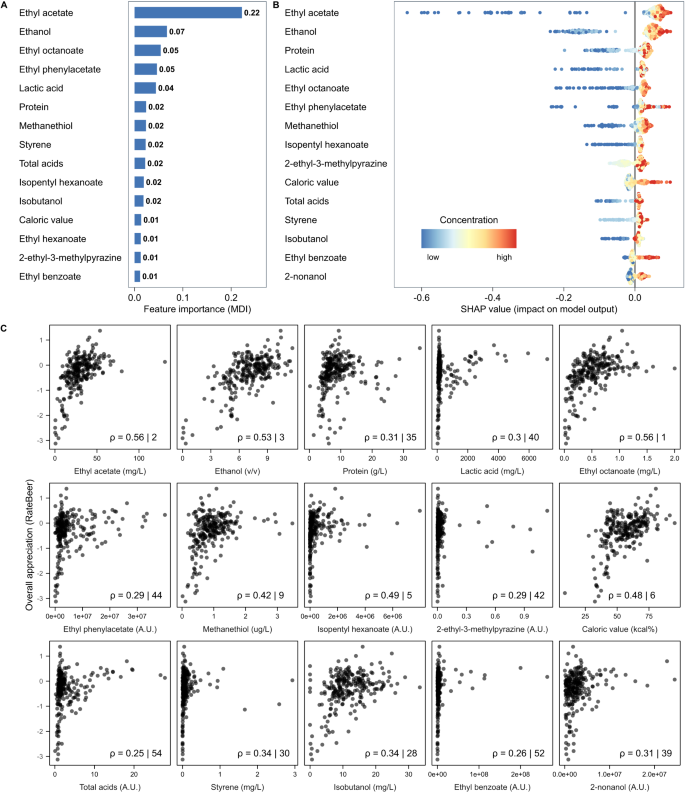
A The impurity-based feature importance (mean deviance in impurity, MDI) calculated from the Gradient Boosting Regression (GBR) model predicting RateBeer appreciation scores. The top 15 highest ranked chemical properties are shown. B SHAP summary plot for the top 15 parameters contributing to our GBR model. Each point on the graph represents a sample from our dataset. The color represents the concentration of that parameter, with bluer colors representing low values and redder colors representing higher values. Greater absolute values on the horizontal axis indicate a higher impact of the parameter on the prediction of the model. C Spearman correlations between the 15 most important chemical properties and consumer overall appreciation. Numbers indicate the Spearman Rho correlation coefficient, and the rank of this correlation compared to all other correlations. The top 15 important compounds were determined using SHAP (panel B).
Both approaches identified ethyl acetate as the most predictive parameter for beer appreciation (Fig. 4 ). Ethyl acetate is the most abundant ester in beer with a typical ‘fruity’, ‘solvent’ and ‘alcoholic’ flavor, but is often considered less important than other esters like isoamyl acetate. The second most important parameter identified by SHAP is ethanol, the most abundant beer compound after water. Apart from directly contributing to beer flavor and mouthfeel, ethanol drastically influences the physical properties of beer, dictating how easily volatile compounds escape the beer matrix to contribute to beer aroma 69 . Importantly, it should also be noted that the importance of ethanol for appreciation is likely inflated by the very low appreciation scores of non-alcoholic beers (Supplementary Fig. S4 ). Despite not often being considered a driver of beer appreciation, protein level also ranks highly in both approaches, possibly due to its effect on mouthfeel and body 70 . Lactic acid, which contributes to the tart taste of sour beers, is the fourth most important parameter identified by SHAP, possibly due to the generally high appreciation of sour beers in our dataset.
Interestingly, some of the most important predictive parameters for our model are not well-established as beer flavors or are even commonly regarded as being negative for beer quality. For example, our models identify methanethiol and ethyl phenyl acetate, an ester commonly linked to beer staling 71 , as a key factor contributing to beer appreciation. Although there is no doubt that high concentrations of these compounds are considered unpleasant, the positive effects of modest concentrations are not yet known 72 , 73 .
To compare our approach to conventional statistics, we evaluated how well the 15 most important SHAP-derived parameters correlate with consumer appreciation (Fig. 4C ). Interestingly, only 6 of the properties derived by SHAP rank amongst the top 15 most correlated parameters. For some chemical compounds, the correlations are so low that they would have likely been considered unimportant. For example, lactic acid, the fourth most important parameter, shows a bimodal distribution for appreciation, with sour beers forming a separate cluster, that is missed entirely by the Spearman correlation. Additionally, the correlation plots reveal outliers, emphasizing the need for robust analysis tools. Together, this highlights the need for alternative models, like the Gradient Boosting model, that better grasp the complexity of (beer) flavor.
Finally, to observe the relationships between these chemical properties and their predicted targets, partial dependence plots were constructed for the six most important predictors of consumer appreciation 74 , 75 , 76 (Supplementary Fig. S7 ). One-way partial dependence plots show how a change in concentration affects the predicted appreciation. These plots reveal an important limitation of our models: appreciation predictions remain constant at ever-increasing concentrations. This implies that once a threshold concentration is reached, further increasing the concentration does not affect appreciation. This is false, as it is well-documented that certain compounds become unpleasant at high concentrations, including ethyl acetate (‘nail polish’) 77 and methanethiol (‘sulfury’ and ‘rotten cabbage’) 78 . The inability of our models to grasp that flavor compounds have optimal levels, above which they become negative, is a consequence of working with commercial beer brands where (off-)flavors are rarely too high to negatively impact the product. The two-way partial dependence plots show how changing the concentration of two compounds influences predicted appreciation, visualizing their interactions (Supplementary Fig. S7 ). In our case, the top 5 parameters are dominated by additive or synergistic interactions, with high concentrations for both compounds resulting in the highest predicted appreciation.
To assess the robustness of our best-performing models and model predictions, we performed 100 iterations of the GBR, RF and ET models. In general, all iterations of the models yielded similar performance (Supplementary Fig. S8 ). Moreover, the main predictors (including the top predictors ethanol and ethyl acetate) remained virtually the same, especially for GBR and RF. For the iterations of the ET model, we did observe more variation in the top predictors, which is likely a consequence of the model’s inherent random architecture in combination with co-correlations between certain predictors. However, even in this case, several of the top predictors (ethanol and ethyl acetate) remain unchanged, although their rank in importance changes (Supplementary Fig. S8 ).
Next, we investigated if a combination of RateBeer and trained panel data into one consolidated dataset would lead to stronger models, under the hypothesis that such a model would suffer less from bias in the datasets. A GBR model was trained to predict appreciation on the combined dataset. This model underperformed compared to the RateBeer model, both in the native case and when including a dataset identifier (R 2 = 0.67, 0.26 and 0.42 respectively). For the latter, the dataset identifier is the most important feature (Supplementary Fig. S9 ), while most of the feature importance remains unchanged, with ethyl acetate and ethanol ranking highest, like in the original model trained only on RateBeer data. It seems that the large variation in the panel dataset introduces noise, weakening the models’ performances and reliability. In addition, it seems reasonable to assume that both datasets are fundamentally different, with the panel dataset obtained by blind tastings by a trained professional panel.
Lastly, we evaluated whether beer style identifiers would further enhance the model’s performance. A GBR model was trained with parameters that explicitly encoded the styles of the samples. This did not improve model performance (R2 = 0.66 with style information vs R2 = 0.67). The most important chemical features are consistent with the model trained without style information (eg. ethanol and ethyl acetate), and with the exception of the most preferred (strong ale) and least preferred (low/no-alcohol) styles, none of the styles were among the most important features (Supplementary Fig. S9 , Supplementary Table S5 and S6 ). This is likely due to a combination of style-specific chemical signatures, such as iso-alpha acids and lactic acid, that implicitly convey style information to the original models, as well as the low number of samples belonging to some styles, making it difficult for the model to learn style-specific patterns. Moreover, beer styles are not rigorously defined, with some styles overlapping in features and some beers being misattributed to a specific style, all of which leads to more noise in models that use style parameters.
Model validation
To test if our predictive models give insight into beer appreciation, we set up experiments aimed at improving existing commercial beers. We specifically selected overall appreciation as the trait to be examined because of its complexity and commercial relevance. Beer flavor comprises a complex bouquet rather than single aromas and tastes 53 . Hence, adding a single compound to the extent that a difference is noticeable may lead to an unbalanced, artificial flavor. Therefore, we evaluated the effect of combinations of compounds. Because Blond beers represent the most extensive style in our dataset, we selected a beer from this style as the starting material for these experiments (Beer 64 in Supplementary Data 1 ).
In the first set of experiments, we adjusted the concentrations of compounds that made up the most important predictors of overall appreciation (ethyl acetate, ethanol, lactic acid, ethyl phenyl acetate) together with correlated compounds (ethyl hexanoate, isoamyl acetate, glycerol), bringing them up to 95 th percentile ethanol-normalized concentrations (Methods) within the Blond group (‘Spiked’ concentration in Fig. 5A ). Compared to controls, the spiked beers were found to have significantly improved overall appreciation among trained panelists, with panelist noting increased intensity of ester flavors, sweetness, alcohol, and body fullness (Fig. 5B ). To disentangle the contribution of ethanol to these results, a second experiment was performed without the addition of ethanol. This resulted in a similar outcome, including increased perception of alcohol and overall appreciation.
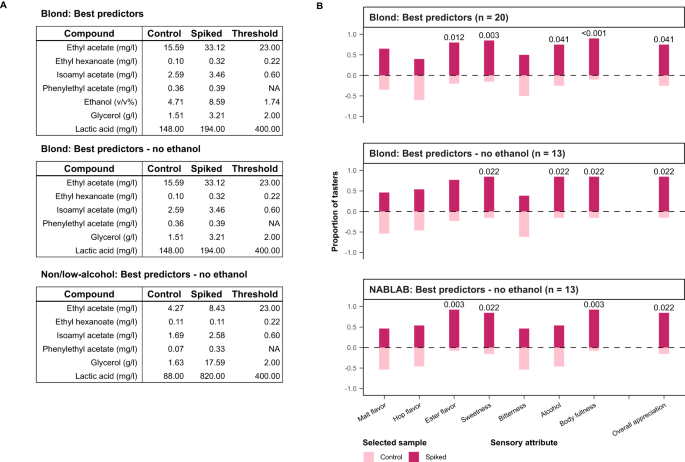
Adding the top chemical compounds, identified as best predictors of appreciation by our model, into poorly appreciated beers results in increased appreciation from our trained panel. Results of sensory tests between base beers and those spiked with compounds identified as the best predictors by the model. A Blond and Non/Low-alcohol (0.0% ABV) base beers were brought up to 95th-percentile ethanol-normalized concentrations within each style. B For each sensory attribute, tasters indicated the more intense sample and selected the sample they preferred. The numbers above the bars correspond to the p values that indicate significant changes in perceived flavor (two-sided binomial test: alpha 0.05, n = 20 or 13).
In a last experiment, we tested whether using the model’s predictions can boost the appreciation of a non-alcoholic beer (beer 223 in Supplementary Data 1 ). Again, the addition of a mixture of predicted compounds (omitting ethanol, in this case) resulted in a significant increase in appreciation, body, ester flavor and sweetness.
Predicting flavor and consumer appreciation from chemical composition is one of the ultimate goals of sensory science. A reliable, systematic and unbiased way to link chemical profiles to flavor and food appreciation would be a significant asset to the food and beverage industry. Such tools would substantially aid in quality control and recipe development, offer an efficient and cost-effective alternative to pilot studies and consumer trials and would ultimately allow food manufacturers to produce superior, tailor-made products that better meet the demands of specific consumer groups more efficiently.
A limited set of studies have previously tried, to varying degrees of success, to predict beer flavor and beer popularity based on (a limited set of) chemical compounds and flavors 79 , 80 . Current sensitive, high-throughput technologies allow measuring an unprecedented number of chemical compounds and properties in a large set of samples, yielding a dataset that can train models that help close the gaps between chemistry and flavor, even for a complex natural product like beer. To our knowledge, no previous research gathered data at this scale (250 samples, 226 chemical parameters, 50 sensory attributes and 5 consumer scores) to disentangle and validate the chemical aspects driving beer preference using various machine-learning techniques. We find that modern machine learning models outperform conventional statistical tools, such as correlations and linear models, and can successfully predict flavor appreciation from chemical composition. This could be attributed to the natural incorporation of interactions and non-linear or discontinuous effects in machine learning models, which are not easily grasped by the linear model architecture. While linear models and partial least squares regression represent the most widespread statistical approaches in sensory science, in part because they allow interpretation 65 , 81 , 82 , modern machine learning methods allow for building better predictive models while preserving the possibility to dissect and exploit the underlying patterns. Of the 10 different models we trained, tree-based models, such as our best performing GBR, showed the best overall performance in predicting sensory responses from chemical information, outcompeting artificial neural networks. This agrees with previous reports for models trained on tabular data 83 . Our results are in line with the findings of Colantonio et al. who also identified the gradient boosting architecture as performing best at predicting appreciation and flavor (of tomatoes and blueberries, in their specific study) 26 . Importantly, besides our larger experimental scale, we were able to directly confirm our models’ predictions in vivo.
Our study confirms that flavor compound concentration does not always correlate with perception, suggesting complex interactions that are often missed by more conventional statistics and simple models. Specifically, we find that tree-based algorithms may perform best in developing models that link complex food chemistry with aroma. Furthermore, we show that massive datasets of untrained consumer reviews provide a valuable source of data, that can complement or even replace trained tasting panels, especially for appreciation and basic flavors, such as sweetness and bitterness. This holds despite biases that are known to occur in such datasets, such as price or conformity bias. Moreover, GBR models predict taste better than aroma. This is likely because taste (e.g. bitterness) often directly relates to the corresponding chemical measurements (e.g., iso-alpha acids), whereas such a link is less clear for aromas, which often result from the interplay between multiple volatile compounds. We also find that our models are best at predicting acidity and alcohol, likely because there is a direct relation between the measured chemical compounds (acids and ethanol) and the corresponding perceived sensorial attribute (acidity and alcohol), and because even untrained consumers are generally able to recognize these flavors and aromas.
The predictions of our final models, trained on review data, hold even for blind tastings with small groups of trained tasters, as demonstrated by our ability to validate specific compounds as drivers of beer flavor and appreciation. Since adding a single compound to the extent of a noticeable difference may result in an unbalanced flavor profile, we specifically tested our identified key drivers as a combination of compounds. While this approach does not allow us to validate if a particular single compound would affect flavor and/or appreciation, our experiments do show that this combination of compounds increases consumer appreciation.
It is important to stress that, while it represents an important step forward, our approach still has several major limitations. A key weakness of the GBR model architecture is that amongst co-correlating variables, the largest main effect is consistently preferred for model building. As a result, co-correlating variables often have artificially low importance scores, both for impurity and SHAP-based methods, like we observed in the comparison to the more randomized Extra Trees models. This implies that chemicals identified as key drivers of a specific sensory feature by GBR might not be the true causative compounds, but rather co-correlate with the actual causative chemical. For example, the high importance of ethyl acetate could be (partially) attributed to the total ester content, ethanol or ethyl hexanoate (rho=0.77, rho=0.72 and rho=0.68), while ethyl phenylacetate could hide the importance of prenyl isobutyrate and ethyl benzoate (rho=0.77 and rho=0.76). Expanding our GBR model to include beer style as a parameter did not yield additional power or insight. This is likely due to style-specific chemical signatures, such as iso-alpha acids and lactic acid, that implicitly convey style information to the original model, as well as the smaller sample size per style, limiting the power to uncover style-specific patterns. This can be partly attributed to the curse of dimensionality, where the high number of parameters results in the models mainly incorporating single parameter effects, rather than complex interactions such as style-dependent effects 67 . A larger number of samples may overcome some of these limitations and offer more insight into style-specific effects. On the other hand, beer style is not a rigid scientific classification, and beers within one style often differ a lot, which further complicates the analysis of style as a model factor.
Our study is limited to beers from Belgian breweries. Although these beers cover a large portion of the beer styles available globally, some beer styles and consumer patterns may be missing, while other features might be overrepresented. For example, many Belgian ales exhibit yeast-driven flavor profiles, which is reflected in the chemical drivers of appreciation discovered by this study. In future work, expanding the scope to include diverse markets and beer styles could lead to the identification of even more drivers of appreciation and better models for special niche products that were not present in our beer set.
In addition to inherent limitations of GBR models, there are also some limitations associated with studying food aroma. Even if our chemical analyses measured most of the known aroma compounds, the total number of flavor compounds in complex foods like beer is still larger than the subset we were able to measure in this study. For example, hop-derived thiols, that influence flavor at very low concentrations, are notoriously difficult to measure in a high-throughput experiment. Moreover, consumer perception remains subjective and prone to biases that are difficult to avoid. It is also important to stress that the models are still immature and that more extensive datasets will be crucial for developing more complete models in the future. Besides more samples and parameters, our dataset does not include any demographic information about the tasters. Including such data could lead to better models that grasp external factors like age and culture. Another limitation is that our set of beers consists of high-quality end-products and lacks beers that are unfit for sale, which limits the current model in accurately predicting products that are appreciated very badly. Finally, while models could be readily applied in quality control, their use in sensory science and product development is restrained by their inability to discern causal relationships. Given that the models cannot distinguish compounds that genuinely drive consumer perception from those that merely correlate, validation experiments are essential to identify true causative compounds.
Despite the inherent limitations, dissection of our models enabled us to pinpoint specific molecules as potential drivers of beer aroma and consumer appreciation, including compounds that were unexpected and would not have been identified using standard approaches. Important drivers of beer appreciation uncovered by our models include protein levels, ethyl acetate, ethyl phenyl acetate and lactic acid. Currently, many brewers already use lactic acid to acidify their brewing water and ensure optimal pH for enzymatic activity during the mashing process. Our results suggest that adding lactic acid can also improve beer appreciation, although its individual effect remains to be tested. Interestingly, ethanol appears to be unnecessary to improve beer appreciation, both for blond beer and alcohol-free beer. Given the growing consumer interest in alcohol-free beer, with a predicted annual market growth of >7% 84 , it is relevant for brewers to know what compounds can further increase consumer appreciation of these beers. Hence, our model may readily provide avenues to further improve the flavor and consumer appreciation of both alcoholic and non-alcoholic beers, which is generally considered one of the key challenges for future beer production.
Whereas we see a direct implementation of our results for the development of superior alcohol-free beverages and other food products, our study can also serve as a stepping stone for the development of novel alcohol-containing beverages. We want to echo the growing body of scientific evidence for the negative effects of alcohol consumption, both on the individual level by the mutagenic, teratogenic and carcinogenic effects of ethanol 85 , 86 , as well as the burden on society caused by alcohol abuse and addiction. We encourage the use of our results for the production of healthier, tastier products, including novel and improved beverages with lower alcohol contents. Furthermore, we strongly discourage the use of these technologies to improve the appreciation or addictive properties of harmful substances.
The present work demonstrates that despite some important remaining hurdles, combining the latest developments in chemical analyses, sensory analysis and modern machine learning methods offers exciting avenues for food chemistry and engineering. Soon, these tools may provide solutions in quality control and recipe development, as well as new approaches to sensory science and flavor research.
Beer selection
250 commercial Belgian beers were selected to cover the broad diversity of beer styles and corresponding diversity in chemical composition and aroma. See Supplementary Fig. S1 .
Chemical dataset
Sample preparation.
Beers within their expiration date were purchased from commercial retailers. Samples were prepared in biological duplicates at room temperature, unless explicitly stated otherwise. Bottle pressure was measured with a manual pressure device (Steinfurth Mess-Systeme GmbH) and used to calculate CO 2 concentration. The beer was poured through two filter papers (Macherey-Nagel, 500713032 MN 713 ¼) to remove carbon dioxide and prevent spontaneous foaming. Samples were then prepared for measurements by targeted Headspace-Gas Chromatography-Flame Ionization Detector/Flame Photometric Detector (HS-GC-FID/FPD), Headspace-Solid Phase Microextraction-Gas Chromatography-Mass Spectrometry (HS-SPME-GC-MS), colorimetric analysis, enzymatic analysis, Near-Infrared (NIR) analysis, as described in the sections below. The mean values of biological duplicates are reported for each compound.
HS-GC-FID/FPD
HS-GC-FID/FPD (Shimadzu GC 2010 Plus) was used to measure higher alcohols, acetaldehyde, esters, 4-vinyl guaicol, and sulfur compounds. Each measurement comprised 5 ml of sample pipetted into a 20 ml glass vial containing 1.75 g NaCl (VWR, 27810.295). 100 µl of 2-heptanol (Sigma-Aldrich, H3003) (internal standard) solution in ethanol (Fisher Chemical, E/0650DF/C17) was added for a final concentration of 2.44 mg/L. Samples were flushed with nitrogen for 10 s, sealed with a silicone septum, stored at −80 °C and analyzed in batches of 20.
The GC was equipped with a DB-WAXetr column (length, 30 m; internal diameter, 0.32 mm; layer thickness, 0.50 µm; Agilent Technologies, Santa Clara, CA, USA) to the FID and an HP-5 column (length, 30 m; internal diameter, 0.25 mm; layer thickness, 0.25 µm; Agilent Technologies, Santa Clara, CA, USA) to the FPD. N 2 was used as the carrier gas. Samples were incubated for 20 min at 70 °C in the headspace autosampler (Flow rate, 35 cm/s; Injection volume, 1000 µL; Injection mode, split; Combi PAL autosampler, CTC analytics, Switzerland). The injector, FID and FPD temperatures were kept at 250 °C. The GC oven temperature was first held at 50 °C for 5 min and then allowed to rise to 80 °C at a rate of 5 °C/min, followed by a second ramp of 4 °C/min until 200 °C kept for 3 min and a final ramp of (4 °C/min) until 230 °C for 1 min. Results were analyzed with the GCSolution software version 2.4 (Shimadzu, Kyoto, Japan). The GC was calibrated with a 5% EtOH solution (VWR International) containing the volatiles under study (Supplementary Table S7 ).
HS-SPME-GC-MS
HS-SPME-GC-MS (Shimadzu GCMS-QP-2010 Ultra) was used to measure additional volatile compounds, mainly comprising terpenoids and esters. Samples were analyzed by HS-SPME using a triphase DVB/Carboxen/PDMS 50/30 μm SPME fiber (Supelco Co., Bellefonte, PA, USA) followed by gas chromatography (Thermo Fisher Scientific Trace 1300 series, USA) coupled to a mass spectrometer (Thermo Fisher Scientific ISQ series MS) equipped with a TriPlus RSH autosampler. 5 ml of degassed beer sample was placed in 20 ml vials containing 1.75 g NaCl (VWR, 27810.295). 5 µl internal standard mix was added, containing 2-heptanol (1 g/L) (Sigma-Aldrich, H3003), 4-fluorobenzaldehyde (1 g/L) (Sigma-Aldrich, 128376), 2,3-hexanedione (1 g/L) (Sigma-Aldrich, 144169) and guaiacol (1 g/L) (Sigma-Aldrich, W253200) in ethanol (Fisher Chemical, E/0650DF/C17). Each sample was incubated at 60 °C in the autosampler oven with constant agitation. After 5 min equilibration, the SPME fiber was exposed to the sample headspace for 30 min. The compounds trapped on the fiber were thermally desorbed in the injection port of the chromatograph by heating the fiber for 15 min at 270 °C.
The GC-MS was equipped with a low polarity RXi-5Sil MS column (length, 20 m; internal diameter, 0.18 mm; layer thickness, 0.18 µm; Restek, Bellefonte, PA, USA). Injection was performed in splitless mode at 320 °C, a split flow of 9 ml/min, a purge flow of 5 ml/min and an open valve time of 3 min. To obtain a pulsed injection, a programmed gas flow was used whereby the helium gas flow was set at 2.7 mL/min for 0.1 min, followed by a decrease in flow of 20 ml/min to the normal 0.9 mL/min. The temperature was first held at 30 °C for 3 min and then allowed to rise to 80 °C at a rate of 7 °C/min, followed by a second ramp of 2 °C/min till 125 °C and a final ramp of 8 °C/min with a final temperature of 270 °C.
Mass acquisition range was 33 to 550 amu at a scan rate of 5 scans/s. Electron impact ionization energy was 70 eV. The interface and ion source were kept at 275 °C and 250 °C, respectively. A mix of linear n-alkanes (from C7 to C40, Supelco Co.) was injected into the GC-MS under identical conditions to serve as external retention index markers. Identification and quantification of the compounds were performed using an in-house developed R script as described in Goelen et al. and Reher et al. 87 , 88 (for package information, see Supplementary Table S8 ). Briefly, chromatograms were analyzed using AMDIS (v2.71) 89 to separate overlapping peaks and obtain pure compound spectra. The NIST MS Search software (v2.0 g) in combination with the NIST2017, FFNSC3 and Adams4 libraries were used to manually identify the empirical spectra, taking into account the expected retention time. After background subtraction and correcting for retention time shifts between samples run on different days based on alkane ladders, compound elution profiles were extracted and integrated using a file with 284 target compounds of interest, which were either recovered in our identified AMDIS list of spectra or were known to occur in beer. Compound elution profiles were estimated for every peak in every chromatogram over a time-restricted window using weighted non-negative least square analysis after which peak areas were integrated 87 , 88 . Batch effect correction was performed by normalizing against the most stable internal standard compound, 4-fluorobenzaldehyde. Out of all 284 target compounds that were analyzed, 167 were visually judged to have reliable elution profiles and were used for final analysis.
Discrete photometric and enzymatic analysis
Discrete photometric and enzymatic analysis (Thermo Scientific TM Gallery TM Plus Beermaster Discrete Analyzer) was used to measure acetic acid, ammonia, beta-glucan, iso-alpha acids, color, sugars, glycerol, iron, pH, protein, and sulfite. 2 ml of sample volume was used for the analyses. Information regarding the reagents and standard solutions used for analyses and calibrations is included in Supplementary Table S7 and Supplementary Table S9 .
NIR analyses
NIR analysis (Anton Paar Alcolyzer Beer ME System) was used to measure ethanol. Measurements comprised 50 ml of sample, and a 10% EtOH solution was used for calibration.
Correlation calculations
Pairwise Spearman Rank correlations were calculated between all chemical properties.
Sensory dataset
Trained panel.
Our trained tasting panel consisted of volunteers who gave prior verbal informed consent. All compounds used for the validation experiment were of food-grade quality. The tasting sessions were approved by the Social and Societal Ethics Committee of the KU Leuven (G-2022-5677-R2(MAR)). All online reviewers agreed to the Terms and Conditions of the RateBeer website.
Sensory analysis was performed according to the American Society of Brewing Chemists (ASBC) Sensory Analysis Methods 90 . 30 volunteers were screened through a series of triangle tests. The sixteen most sensitive and consistent tasters were retained as taste panel members. The resulting panel was diverse in age [22–42, mean: 29], sex [56% male] and nationality [7 different countries]. The panel developed a consensus vocabulary to describe beer aroma, taste and mouthfeel. Panelists were trained to identify and score 50 different attributes, using a 7-point scale to rate attributes’ intensity. The scoring sheet is included as Supplementary Data 3 . Sensory assessments took place between 10–12 a.m. The beers were served in black-colored glasses. Per session, between 5 and 12 beers of the same style were tasted at 12 °C to 16 °C. Two reference beers were added to each set and indicated as ‘Reference 1 & 2’, allowing panel members to calibrate their ratings. Not all panelists were present at every tasting. Scores were scaled by standard deviation and mean-centered per taster. Values are represented as z-scores and clustered by Euclidean distance. Pairwise Spearman correlations were calculated between taste and aroma sensory attributes. Panel consistency was evaluated by repeating samples on different sessions and performing ANOVA to identify differences, using the ‘stats’ package (v4.2.2) in R (for package information, see Supplementary Table S8 ).
Online reviews from a public database
The ‘scrapy’ package in Python (v3.6) (for package information, see Supplementary Table S8 ). was used to collect 232,288 online reviews (mean=922, min=6, max=5343) from RateBeer, an online beer review database. Each review entry comprised 5 numerical scores (appearance, aroma, taste, palate and overall quality) and an optional review text. The total number of reviews per reviewer was collected separately. Numerical scores were scaled and centered per rater, and mean scores were calculated per beer.
For the review texts, the language was estimated using the packages ‘langdetect’ and ‘langid’ in Python. Reviews that were classified as English by both packages were kept. Reviewers with fewer than 100 entries overall were discarded. 181,025 reviews from >6000 reviewers from >40 countries remained. Text processing was done using the ‘nltk’ package in Python. Texts were corrected for slang and misspellings; proper nouns and rare words that are relevant to the beer context were specified and kept as-is (‘Chimay’,’Lambic’, etc.). A dictionary of semantically similar sensorial terms, for example ‘floral’ and ‘flower’, was created and collapsed together into one term. Words were stemmed and lemmatized to avoid identifying words such as ‘acid’ and ‘acidity’ as separate terms. Numbers and punctuation were removed.
Sentences from up to 50 randomly chosen reviews per beer were manually categorized according to the aspect of beer they describe (appearance, aroma, taste, palate, overall quality—not to be confused with the 5 numerical scores described above) or flagged as irrelevant if they contained no useful information. If a beer contained fewer than 50 reviews, all reviews were manually classified. This labeled data set was used to train a model that classified the rest of the sentences for all beers 91 . Sentences describing taste and aroma were extracted, and term frequency–inverse document frequency (TFIDF) was implemented to calculate enrichment scores for sensorial words per beer.
The sex of the tasting subject was not considered when building our sensory database. Instead, results from different panelists were averaged, both for our trained panel (56% male, 44% female) and the RateBeer reviews (70% male, 30% female for RateBeer as a whole).
Beer price collection and processing
Beer prices were collected from the following stores: Colruyt, Delhaize, Total Wine, BeerHawk, The Belgian Beer Shop, The Belgian Shop, and Beer of Belgium. Where applicable, prices were converted to Euros and normalized per liter. Spearman correlations were calculated between these prices and mean overall appreciation scores from RateBeer and the taste panel, respectively.
Pairwise Spearman Rank correlations were calculated between all sensory properties.
Machine learning models
Predictive modeling of sensory profiles from chemical data.
Regression models were constructed to predict (a) trained panel scores for beer flavors and quality from beer chemical profiles and (b) public reviews’ appreciation scores from beer chemical profiles. Z-scores were used to represent sensory attributes in both data sets. Chemical properties with log-normal distributions (Shapiro-Wilk test, p < 0.05 ) were log-transformed. Missing chemical measurements (0.1% of all data) were replaced with mean values per attribute. Observations from 250 beers were randomly separated into a training set (70%, 175 beers) and a test set (30%, 75 beers), stratified per beer style. Chemical measurements (p = 231) were normalized based on the training set average and standard deviation. In total, three linear regression-based models: linear regression with first-order interaction terms (LR), lasso regression with first-order interaction terms (Lasso) and partial least squares regression (PLSR); five decision tree models, Adaboost regressor (ABR), Extra Trees (ET), Gradient Boosting regressor (GBR), Random Forest (RF) and XGBoost regressor (XGBR); one support vector machine model (SVR) and one artificial neural network model (ANN) were trained. The models were implemented using the ‘scikit-learn’ package (v1.2.2) and ‘xgboost’ package (v1.7.3) in Python (v3.9.16). Models were trained, and hyperparameters optimized, using five-fold cross-validated grid search with the coefficient of determination (R 2 ) as the evaluation metric. The ANN (scikit-learn’s MLPRegressor) was optimized using Bayesian Tree-Structured Parzen Estimator optimization with the ‘Optuna’ Python package (v3.2.0). Individual models were trained per attribute, and a multi-output model was trained on all attributes simultaneously.
Model dissection
GBR was found to outperform other methods, resulting in models with the highest average R 2 values in both trained panel and public review data sets. Impurity-based rankings of the most important predictors for each predicted sensorial trait were obtained using the ‘scikit-learn’ package. To observe the relationships between these chemical properties and their predicted targets, partial dependence plots (PDP) were constructed for the six most important predictors of consumer appreciation 74 , 75 .
The ‘SHAP’ package in Python (v0.41.0) was implemented to provide an alternative ranking of predictor importance and to visualize the predictors’ effects as a function of their concentration 68 .
Validation of causal chemical properties
To validate the effects of the most important model features on predicted sensory attributes, beers were spiked with the chemical compounds identified by the models and descriptive sensory analyses were carried out according to the American Society of Brewing Chemists (ASBC) protocol 90 .
Compound spiking was done 30 min before tasting. Compounds were spiked into fresh beer bottles, that were immediately resealed and inverted three times. Fresh bottles of beer were opened for the same duration, resealed, and inverted thrice, to serve as controls. Pairs of spiked samples and controls were served simultaneously, chilled and in dark glasses as outlined in the Trained panel section above. Tasters were instructed to select the glass with the higher flavor intensity for each attribute (directional difference test 92 ) and to select the glass they prefer.
The final concentration after spiking was equal to the within-style average, after normalizing by ethanol concentration. This was done to ensure balanced flavor profiles in the final spiked beer. The same methods were applied to improve a non-alcoholic beer. Compounds were the following: ethyl acetate (Merck KGaA, W241415), ethyl hexanoate (Merck KGaA, W243906), isoamyl acetate (Merck KGaA, W205508), phenethyl acetate (Merck KGaA, W285706), ethanol (96%, Colruyt), glycerol (Merck KGaA, W252506), lactic acid (Merck KGaA, 261106).
Significant differences in preference or perceived intensity were determined by performing the two-sided binomial test on each attribute.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data that support the findings of this work are available in the Supplementary Data files and have been deposited to Zenodo under accession code 10653704 93 . The RateBeer scores data are under restricted access, they are not publicly available as they are property of RateBeer (ZX Ventures, USA). Access can be obtained from the authors upon reasonable request and with permission of RateBeer (ZX Ventures, USA). Source data are provided with this paper.
Code availability
The code for training the machine learning models, analyzing the models, and generating the figures has been deposited to Zenodo under accession code 10653704 93 .
Tieman, D. et al. A chemical genetic roadmap to improved tomato flavor. Science 355 , 391–394 (2017).
Article ADS CAS PubMed Google Scholar
Plutowska, B. & Wardencki, W. Application of gas chromatography–olfactometry (GC–O) in analysis and quality assessment of alcoholic beverages – A review. Food Chem. 107 , 449–463 (2008).
Article CAS Google Scholar
Legin, A., Rudnitskaya, A., Seleznev, B. & Vlasov, Y. Electronic tongue for quality assessment of ethanol, vodka and eau-de-vie. Anal. Chim. Acta 534 , 129–135 (2005).
Loutfi, A., Coradeschi, S., Mani, G. K., Shankar, P. & Rayappan, J. B. B. Electronic noses for food quality: A review. J. Food Eng. 144 , 103–111 (2015).
Ahn, Y.-Y., Ahnert, S. E., Bagrow, J. P. & Barabási, A.-L. Flavor network and the principles of food pairing. Sci. Rep. 1 , 196 (2011).
Article CAS PubMed PubMed Central Google Scholar
Bartoshuk, L. M. & Klee, H. J. Better fruits and vegetables through sensory analysis. Curr. Biol. 23 , R374–R378 (2013).
Article CAS PubMed Google Scholar
Piggott, J. R. Design questions in sensory and consumer science. Food Qual. Prefer. 3293 , 217–220 (1995).
Article Google Scholar
Kermit, M. & Lengard, V. Assessing the performance of a sensory panel-panellist monitoring and tracking. J. Chemom. 19 , 154–161 (2005).
Cook, D. J., Hollowood, T. A., Linforth, R. S. T. & Taylor, A. J. Correlating instrumental measurements of texture and flavour release with human perception. Int. J. Food Sci. Technol. 40 , 631–641 (2005).
Chinchanachokchai, S., Thontirawong, P. & Chinchanachokchai, P. A tale of two recommender systems: The moderating role of consumer expertise on artificial intelligence based product recommendations. J. Retail. Consum. Serv. 61 , 1–12 (2021).
Ross, C. F. Sensory science at the human-machine interface. Trends Food Sci. Technol. 20 , 63–72 (2009).
Chambers, E. IV & Koppel, K. Associations of volatile compounds with sensory aroma and flavor: The complex nature of flavor. Molecules 18 , 4887–4905 (2013).
Pinu, F. R. Metabolomics—The new frontier in food safety and quality research. Food Res. Int. 72 , 80–81 (2015).
Danezis, G. P., Tsagkaris, A. S., Brusic, V. & Georgiou, C. A. Food authentication: state of the art and prospects. Curr. Opin. Food Sci. 10 , 22–31 (2016).
Shepherd, G. M. Smell images and the flavour system in the human brain. Nature 444 , 316–321 (2006).
Meilgaard, M. C. Prediction of flavor differences between beers from their chemical composition. J. Agric. Food Chem. 30 , 1009–1017 (1982).
Xu, L. et al. Widespread receptor-driven modulation in peripheral olfactory coding. Science 368 , eaaz5390 (2020).
Kupferschmidt, K. Following the flavor. Science 340 , 808–809 (2013).
Billesbølle, C. B. et al. Structural basis of odorant recognition by a human odorant receptor. Nature 615 , 742–749 (2023).
Article ADS PubMed PubMed Central Google Scholar
Smith, B. Perspective: Complexities of flavour. Nature 486 , S6–S6 (2012).
Pfister, P. et al. Odorant receptor inhibition is fundamental to odor encoding. Curr. Biol. 30 , 2574–2587 (2020).
Moskowitz, H. W., Kumaraiah, V., Sharma, K. N., Jacobs, H. L. & Sharma, S. D. Cross-cultural differences in simple taste preferences. Science 190 , 1217–1218 (1975).
Eriksson, N. et al. A genetic variant near olfactory receptor genes influences cilantro preference. Flavour 1 , 22 (2012).
Ferdenzi, C. et al. Variability of affective responses to odors: Culture, gender, and olfactory knowledge. Chem. Senses 38 , 175–186 (2013).
Article PubMed Google Scholar
Lawless, H. T. & Heymann, H. Sensory evaluation of food: Principles and practices. (Springer, New York, NY). https://doi.org/10.1007/978-1-4419-6488-5 (2010).
Colantonio, V. et al. Metabolomic selection for enhanced fruit flavor. Proc. Natl. Acad. Sci. 119 , e2115865119 (2022).
Fritz, F., Preissner, R. & Banerjee, P. VirtualTaste: a web server for the prediction of organoleptic properties of chemical compounds. Nucleic Acids Res 49 , W679–W684 (2021).
Tuwani, R., Wadhwa, S. & Bagler, G. BitterSweet: Building machine learning models for predicting the bitter and sweet taste of small molecules. Sci. Rep. 9 , 1–13 (2019).
Dagan-Wiener, A. et al. Bitter or not? BitterPredict, a tool for predicting taste from chemical structure. Sci. Rep. 7 , 1–13 (2017).
Pallante, L. et al. Toward a general and interpretable umami taste predictor using a multi-objective machine learning approach. Sci. Rep. 12 , 1–11 (2022).
Malavolta, M. et al. A survey on computational taste predictors. Eur. Food Res. Technol. 248 , 2215–2235 (2022).
Lee, B. K. et al. A principal odor map unifies diverse tasks in olfactory perception. Science 381 , 999–1006 (2023).
Mayhew, E. J. et al. Transport features predict if a molecule is odorous. Proc. Natl. Acad. Sci. 119 , e2116576119 (2022).
Niu, Y. et al. Sensory evaluation of the synergism among ester odorants in light aroma-type liquor by odor threshold, aroma intensity and flash GC electronic nose. Food Res. Int. 113 , 102–114 (2018).
Yu, P., Low, M. Y. & Zhou, W. Design of experiments and regression modelling in food flavour and sensory analysis: A review. Trends Food Sci. Technol. 71 , 202–215 (2018).
Oladokun, O. et al. The impact of hop bitter acid and polyphenol profiles on the perceived bitterness of beer. Food Chem. 205 , 212–220 (2016).
Linforth, R., Cabannes, M., Hewson, L., Yang, N. & Taylor, A. Effect of fat content on flavor delivery during consumption: An in vivo model. J. Agric. Food Chem. 58 , 6905–6911 (2010).
Guo, S., Na Jom, K. & Ge, Y. Influence of roasting condition on flavor profile of sunflower seeds: A flavoromics approach. Sci. Rep. 9 , 11295 (2019).
Ren, Q. et al. The changes of microbial community and flavor compound in the fermentation process of Chinese rice wine using Fagopyrum tataricum grain as feedstock. Sci. Rep. 9 , 3365 (2019).
Hastie, T., Friedman, J. & Tibshirani, R. The Elements of Statistical Learning. (Springer, New York, NY). https://doi.org/10.1007/978-0-387-21606-5 (2001).
Dietz, C., Cook, D., Huismann, M., Wilson, C. & Ford, R. The multisensory perception of hop essential oil: a review. J. Inst. Brew. 126 , 320–342 (2020).
CAS Google Scholar
Roncoroni, Miguel & Verstrepen, Kevin Joan. Belgian Beer: Tested and Tasted. (Lannoo, 2018).
Meilgaard, M. Flavor chemistry of beer: Part II: Flavor and threshold of 239 aroma volatiles. in (1975).
Bokulich, N. A. & Bamforth, C. W. The microbiology of malting and brewing. Microbiol. Mol. Biol. Rev. MMBR 77 , 157–172 (2013).
Dzialo, M. C., Park, R., Steensels, J., Lievens, B. & Verstrepen, K. J. Physiology, ecology and industrial applications of aroma formation in yeast. FEMS Microbiol. Rev. 41 , S95–S128 (2017).
Article PubMed PubMed Central Google Scholar
Datta, A. et al. Computer-aided food engineering. Nat. Food 3 , 894–904 (2022).
American Society of Brewing Chemists. Beer Methods. (American Society of Brewing Chemists, St. Paul, MN, U.S.A.).
Olaniran, A. O., Hiralal, L., Mokoena, M. P. & Pillay, B. Flavour-active volatile compounds in beer: production, regulation and control. J. Inst. Brew. 123 , 13–23 (2017).
Verstrepen, K. J. et al. Flavor-active esters: Adding fruitiness to beer. J. Biosci. Bioeng. 96 , 110–118 (2003).
Meilgaard, M. C. Flavour chemistry of beer. part I: flavour interaction between principal volatiles. Master Brew. Assoc. Am. Tech. Q 12 , 107–117 (1975).
Briggs, D. E., Boulton, C. A., Brookes, P. A. & Stevens, R. Brewing 227–254. (Woodhead Publishing). https://doi.org/10.1533/9781855739062.227 (2004).
Bossaert, S., Crauwels, S., De Rouck, G. & Lievens, B. The power of sour - A review: Old traditions, new opportunities. BrewingScience 72 , 78–88 (2019).
Google Scholar
Verstrepen, K. J. et al. Flavor active esters: Adding fruitiness to beer. J. Biosci. Bioeng. 96 , 110–118 (2003).
Snauwaert, I. et al. Microbial diversity and metabolite composition of Belgian red-brown acidic ales. Int. J. Food Microbiol. 221 , 1–11 (2016).
Spitaels, F. et al. The microbial diversity of traditional spontaneously fermented lambic beer. PLoS ONE 9 , e95384 (2014).
Blanco, C. A., Andrés-Iglesias, C. & Montero, O. Low-alcohol Beers: Flavor Compounds, Defects, and Improvement Strategies. Crit. Rev. Food Sci. Nutr. 56 , 1379–1388 (2016).
Jackowski, M. & Trusek, A. Non-Alcohol. beer Prod. – Overv. 20 , 32–38 (2018).
Takoi, K. et al. The contribution of geraniol metabolism to the citrus flavour of beer: Synergy of geraniol and β-citronellol under coexistence with excess linalool. J. Inst. Brew. 116 , 251–260 (2010).
Kroeze, J. H. & Bartoshuk, L. M. Bitterness suppression as revealed by split-tongue taste stimulation in humans. Physiol. Behav. 35 , 779–783 (1985).
Mennella, J. A. et al. A spoonful of sugar helps the medicine go down”: Bitter masking bysucrose among children and adults. Chem. Senses 40 , 17–25 (2015).
Wietstock, P., Kunz, T., Perreira, F. & Methner, F.-J. Metal chelation behavior of hop acids in buffered model systems. BrewingScience 69 , 56–63 (2016).
Sancho, D., Blanco, C. A., Caballero, I. & Pascual, A. Free iron in pale, dark and alcohol-free commercial lager beers. J. Sci. Food Agric. 91 , 1142–1147 (2011).
Rodrigues, H. & Parr, W. V. Contribution of cross-cultural studies to understanding wine appreciation: A review. Food Res. Int. 115 , 251–258 (2019).
Korneva, E. & Blockeel, H. Towards better evaluation of multi-target regression models. in ECML PKDD 2020 Workshops (eds. Koprinska, I. et al.) 353–362 (Springer International Publishing, Cham, 2020). https://doi.org/10.1007/978-3-030-65965-3_23 .
Gastón Ares. Mathematical and Statistical Methods in Food Science and Technology. (Wiley, 2013).
Grinsztajn, L., Oyallon, E. & Varoquaux, G. Why do tree-based models still outperform deep learning on tabular data? Preprint at http://arxiv.org/abs/2207.08815 (2022).
Gries, S. T. Statistics for Linguistics with R: A Practical Introduction. in Statistics for Linguistics with R (De Gruyter Mouton, 2021). https://doi.org/10.1515/9783110718256 .
Lundberg, S. M. et al. From local explanations to global understanding with explainable AI for trees. Nat. Mach. Intell. 2 , 56–67 (2020).
Ickes, C. M. & Cadwallader, K. R. Effects of ethanol on flavor perception in alcoholic beverages. Chemosens. Percept. 10 , 119–134 (2017).
Kato, M. et al. Influence of high molecular weight polypeptides on the mouthfeel of commercial beer. J. Inst. Brew. 127 , 27–40 (2021).
Wauters, R. et al. Novel Saccharomyces cerevisiae variants slow down the accumulation of staling aldehydes and improve beer shelf-life. Food Chem. 398 , 1–11 (2023).
Li, H., Jia, S. & Zhang, W. Rapid determination of low-level sulfur compounds in beer by headspace gas chromatography with a pulsed flame photometric detector. J. Am. Soc. Brew. Chem. 66 , 188–191 (2008).
Dercksen, A., Laurens, J., Torline, P., Axcell, B. C. & Rohwer, E. Quantitative analysis of volatile sulfur compounds in beer using a membrane extraction interface. J. Am. Soc. Brew. Chem. 54 , 228–233 (1996).
Molnar, C. Interpretable Machine Learning: A Guide for Making Black-Box Models Interpretable. (2020).
Zhao, Q. & Hastie, T. Causal interpretations of black-box models. J. Bus. Econ. Stat. Publ. Am. Stat. Assoc. 39 , 272–281 (2019).
Article MathSciNet Google Scholar
Hastie, T., Tibshirani, R. & Friedman, J. The Elements of Statistical Learning. (Springer, 2019).
Labrado, D. et al. Identification by NMR of key compounds present in beer distillates and residual phases after dealcoholization by vacuum distillation. J. Sci. Food Agric. 100 , 3971–3978 (2020).
Lusk, L. T., Kay, S. B., Porubcan, A. & Ryder, D. S. Key olfactory cues for beer oxidation. J. Am. Soc. Brew. Chem. 70 , 257–261 (2012).
Gonzalez Viejo, C., Torrico, D. D., Dunshea, F. R. & Fuentes, S. Development of artificial neural network models to assess beer acceptability based on sensory properties using a robotic pourer: A comparative model approach to achieve an artificial intelligence system. Beverages 5 , 33 (2019).
Gonzalez Viejo, C., Fuentes, S., Torrico, D. D., Godbole, A. & Dunshea, F. R. Chemical characterization of aromas in beer and their effect on consumers liking. Food Chem. 293 , 479–485 (2019).
Gilbert, J. L. et al. Identifying breeding priorities for blueberry flavor using biochemical, sensory, and genotype by environment analyses. PLOS ONE 10 , 1–21 (2015).
Goulet, C. et al. Role of an esterase in flavor volatile variation within the tomato clade. Proc. Natl. Acad. Sci. 109 , 19009–19014 (2012).
Article ADS CAS PubMed PubMed Central Google Scholar
Borisov, V. et al. Deep Neural Networks and Tabular Data: A Survey. IEEE Trans. Neural Netw. Learn. Syst. 1–21 https://doi.org/10.1109/TNNLS.2022.3229161 (2022).
Statista. Statista Consumer Market Outlook: Beer - Worldwide.
Seitz, H. K. & Stickel, F. Molecular mechanisms of alcoholmediated carcinogenesis. Nat. Rev. Cancer 7 , 599–612 (2007).
Voordeckers, K. et al. Ethanol exposure increases mutation rate through error-prone polymerases. Nat. Commun. 11 , 3664 (2020).
Goelen, T. et al. Bacterial phylogeny predicts volatile organic compound composition and olfactory response of an aphid parasitoid. Oikos 129 , 1415–1428 (2020).
Article ADS Google Scholar
Reher, T. et al. Evaluation of hop (Humulus lupulus) as a repellent for the management of Drosophila suzukii. Crop Prot. 124 , 104839 (2019).
Stein, S. E. An integrated method for spectrum extraction and compound identification from gas chromatography/mass spectrometry data. J. Am. Soc. Mass Spectrom. 10 , 770–781 (1999).
American Society of Brewing Chemists. Sensory Analysis Methods. (American Society of Brewing Chemists, St. Paul, MN, U.S.A., 1992).
McAuley, J., Leskovec, J. & Jurafsky, D. Learning Attitudes and Attributes from Multi-Aspect Reviews. Preprint at https://doi.org/10.48550/arXiv.1210.3926 (2012).
Meilgaard, M. C., Carr, B. T. & Carr, B. T. Sensory Evaluation Techniques. (CRC Press, Boca Raton). https://doi.org/10.1201/b16452 (2014).
Schreurs, M. et al. Data from: Predicting and improving complex beer flavor through machine learning. Zenodo https://doi.org/10.5281/zenodo.10653704 (2024).
Download references
Acknowledgements
We thank all lab members for their discussions and thank all tasting panel members for their contributions. Special thanks go out to Dr. Karin Voordeckers for her tremendous help in proofreading and improving the manuscript. M.S. was supported by a Baillet-Latour fellowship, L.C. acknowledges financial support from KU Leuven (C16/17/006), F.A.T. was supported by a PhD fellowship from FWO (1S08821N). Research in the lab of K.J.V. is supported by KU Leuven, FWO, VIB, VLAIO and the Brewing Science Serves Health Fund. Research in the lab of T.W. is supported by FWO (G.0A51.15) and KU Leuven (C16/17/006).
Author information
These authors contributed equally: Michiel Schreurs, Supinya Piampongsant, Miguel Roncoroni.
Authors and Affiliations
VIB—KU Leuven Center for Microbiology, Gaston Geenslaan 1, B-3001, Leuven, Belgium
Michiel Schreurs, Supinya Piampongsant, Miguel Roncoroni, Lloyd Cool, Beatriz Herrera-Malaver, Florian A. Theßeling & Kevin J. Verstrepen
CMPG Laboratory of Genetics and Genomics, KU Leuven, Gaston Geenslaan 1, B-3001, Leuven, Belgium
Leuven Institute for Beer Research (LIBR), Gaston Geenslaan 1, B-3001, Leuven, Belgium
Laboratory of Socioecology and Social Evolution, KU Leuven, Naamsestraat 59, B-3000, Leuven, Belgium
Lloyd Cool, Christophe Vanderaa & Tom Wenseleers
VIB Bioinformatics Core, VIB, Rijvisschestraat 120, B-9052, Ghent, Belgium
Łukasz Kreft & Alexander Botzki
AB InBev SA/NV, Brouwerijplein 1, B-3000, Leuven, Belgium
Philippe Malcorps & Luk Daenen
You can also search for this author in PubMed Google Scholar
Contributions
S.P., M.S. and K.J.V. conceived the experiments. S.P., M.S. and K.J.V. designed the experiments. S.P., M.S., M.R., B.H. and F.A.T. performed the experiments. S.P., M.S., L.C., C.V., L.K., A.B., P.M., L.D., T.W. and K.J.V. contributed analysis ideas. S.P., M.S., L.C., C.V., T.W. and K.J.V. analyzed the data. All authors contributed to writing the manuscript.
Corresponding author
Correspondence to Kevin J. Verstrepen .
Ethics declarations
Competing interests.
K.J.V. is affiliated with bar.on. The other authors declare no competing interests.
Peer review
Peer review information.
Nature Communications thanks Florian Bauer, Andrew John Macintosh and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information, peer review file, description of additional supplementary files, supplementary data 1, supplementary data 2, supplementary data 3, supplementary data 4, supplementary data 5, supplementary data 6, supplementary data 7, reporting summary, source data, source data, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Schreurs, M., Piampongsant, S., Roncoroni, M. et al. Predicting and improving complex beer flavor through machine learning. Nat Commun 15 , 2368 (2024). https://doi.org/10.1038/s41467-024-46346-0
Download citation
Received : 30 October 2023
Accepted : 21 February 2024
Published : 26 March 2024
DOI : https://doi.org/10.1038/s41467-024-46346-0
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
By submitting a comment you agree to abide by our Terms and Community Guidelines . If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing: Translational Research newsletter — top stories in biotechnology, drug discovery and pharma.
- Quantum Research
Landmark IBM error correction paper published on the cover of Nature
Ibm has created a quantum error-correcting code about 10 times more efficient than prior methods — a milestone in quantum computing research..

27 Mar 2024
Rafi Letzter
Share this blog
Today, the paper detailing those results was published as the cover story of the scientific journal Nature. 1
Last year, we demonstrated that quantum computers had entered the era of utility , where they are now capable of running quantum circuits better than classical computers can. Over the next few years, we expect to find speedups over classical computing and extract business value from these systems. But there are also algorithms with mathematically proven speedups over leading classical methods that require tuning quantum circuits with hundreds of millions, to billions, of gates. Expanding our quantum computing toolkit to include those algorithms requires us to find a way to compute that corrects the errors inherent to quantum systems — what we call quantum error correction.
Read how a paper from IBM and UC Berkeley shows a path toward useful quantum computing
Quantum error correction requires that we encode quantum information into more qubits than we would otherwise need. However, achieving quantum error correction in a scalable and fault-tolerant way has, to this point, been out of reach without considering scales of one million or more physical qubits. Our new result published today greatly reduces that overhead, and shows that error correction is within reach.
While quantum error correction theory dates back three decades, theoretical error correction techniques capable of running valuable quantum circuits on real hardware have been too impractical to deploy on quantum system. In our new paper, we introduce a new code, which we call the gross code , that overcomes that limitation.
This code is part of our broader strategy to bring useful quantum computing to the world.
While error correction is not a solved problem, this new code makes clear the path toward running quantum circuits with a billion gates or more on our superconducting transmon qubit hardware.
What is error correction?
Quantum information is fragile and susceptible to noise — environmental noise, noise from the control electronics, hardware imperfections, state preparation and measurement errors, and more. In order to run quantum circuits with millions to billions of gates, quantum error correction will be required.
Error correction works by building redundancy into quantum circuits. Many qubits work together to protect a piece of quantum information that a single qubit might lose to errors and noise.
On classical computers, the concept of redundancy is pretty straightforward. Classical error correction involves storing the same piece of information across multiple bits. Instead of storing a 1 as a 1 or a 0 as a 0, the computer might record 11111 or 00000. That way, if an error flips a minority of bits, the computer can treat 11001 as 1, or 10001 as 0. It’s fairly easy to build in more redundancy as needed to introduce finer error correction.
Things are more complicated on quantum computers. Quantum information cannot be copied and pasted like classical information, and the information stored in quantum bits is more complicated than classical data. And of course, qubits can decohere quickly, forgetting their stored information.
Research has shown that quantum fault tolerance is possible, and there are many error correcting schemes on the books. The most popular one is called the “surface code,” where qubits are arranged on a two-dimensional lattice and units of information are encoded into sub-units of the lattice.
But these schemes have problems.
First, they only work if the hardware’s error rates are better than some threshold determined by the specific scheme and the properties of the noise itself — and beating those thresholds can be a challenge.
Second, many of those schemes scale inefficiently — as you build larger quantum computers, the number of extra qubits needed for error correction far outpaces the number of qubits the code can store.
At practical code sizes where many errors can be corrected, the surface code uses hundreds of physical qubits per encoded qubit worth of quantum information, or more. So, while the surface code is useful for benchmarking and learning about error correction, it’s probably not the end of the story for fault-tolerant quantum computers.
Exploring “good” codes
The field of error correction buzzed with excitement in 2022 when Pavel Panteleev and Gleb Kalachev at Moscow State University published a landmark paper proving that there exist asymptotically good codes — codes where the number of extra qubits needed levels off as the quality of the code increases.
This has spurred a lot of new work in error correction, especially in the same family of codes that the surface code hails from, called quantum low-density parity check, or qLDPC codes. These qLDPC codes are quantum error correcting codes where the operations responsible for checking whether or not an error has occurred only have to act on a few qubits, and each qubit only has to participate in a few checks.
But this work was highly theoretical, focused on proving the possibility of this kind of error correction. It didn’t take into account the real constraints of building quantum computers. Most importantly, some qLDPC codes would require many qubits in a system to be physically linked to high numbers of other qubits. In practice, that would require quantum processors folded in on themselves in psychedelic hyper-dimensional origami, or entombed in wildly complex rats’ nests of wires.
In our paper, we looked for fault-tolerant quantum memory with a low qubit overhead, high error threshold, and a large code distance.

Bravyi, S., Cross, A., Gambetta, J., et al. High-threshold and low-overhead fault-tolerant quantum memory. Nature (2024). https://doi.org/10.1038/s41586-024-07107-7
In our Nature paper, we specifically looked for fault-tolerant quantum memory with a low qubit overhead, high error threshold, and a large code distance.
Let’s break that down:
Fault-tolerant: The circuits used to detect errors won't spread those errors around too badly in the process, and they can be corrected faster than they occur
Quantum memory: In this paper, we are only encoding and storing quantum information. We are not yet doing calculations on the encoded quantum information.
High error threshold: The higher the threshold, the higher amount of hardware errors the code will allow while still being fault tolerant. We were looking for a code that allowed us to operate the memory reliably at physical error rates as high as 0.001, so we wanted a threshold close to 1 percent.
Large code distance: Distance is the measure of how robust the code is — how many errors it takes to completely flip the value from 0 to 1 and vice versa. In the case of 00000 and 11111, the distance is 5. We wanted one with a large code distance that corrects more than just a couple errors. Large-distance codes can suppress noise by orders of magnitude even if the hardware quality is only marginally better than the code threshold. In contrast, codes with a small distance become useful only if the hardware quality is significantly better than the code threshold.
Low qubit overhead: Overhead is the number of extra qubits required for correcting errors. We want the number of qubits required to do error correction to be far less than we need for a surface code of the same quality, or distance.
We’re excited to report that our team’s mathematical analysis found concrete examples of qLDPC codes that met all of these required conditions. These fall into a family of codes called “Bivariate Bicycle (BB)” codes. And they are going to shape not only our research going forward, but how we architect physical quantum systems.
The gross code
While many qLDPC code families show great promise for advancing error correction theory, most aren’t necessarily pragmatic for real-world application. Our new codes lend themselves better to practical implementation because each qubit needs only to connect to six others, and the connections can be routed on just two layers.
To get an idea of how the qubits are connected, imagine they are put onto a square grid, like a piece of graph paper. Curl up this piece of graph paper so that it forms a tube, and connect the ends of the tube to make a donut. On this donut, each qubit is connected to its four neighbors and two qubits that are farther away on the surface of the donut. No more connections needed.
The good news is we don’t actually have to embed our qubits onto a donut to make these codes work — we can accomplish this by folding the surface differently and adding a few other long-range connectors to satisfy mathematical requirements of the code. It’s an engineering challenge, but much more feasible than a hyper-dimensional shape.
We explored some codes that have this architecture and focused on a particular [[144,12,12]] code. We call this code the gross code because 144 is a gross (or a dozen dozen). It requires 144 qubits to store data — but in our specific implementation, it also uses another 144 qubits to check for errors, so this instance of the code uses 288 qubits. It stores 12 logical qubits well enough that fewer than 12 errors can be detected. Thus: [[144,12,12]].
Using the gross code, you can protect 12 logical qubits for roughly a million cycles of error checks using 288 qubits. Doing roughly the same task with the surface code would require nearly 3,000 qubits.
This is a milestone. We are still looking for qLDPC codes with even more efficient architectures, and our research on performing error-corrected calculations using these codes is ongoing. But with this publication, the future of error correction looks bright.
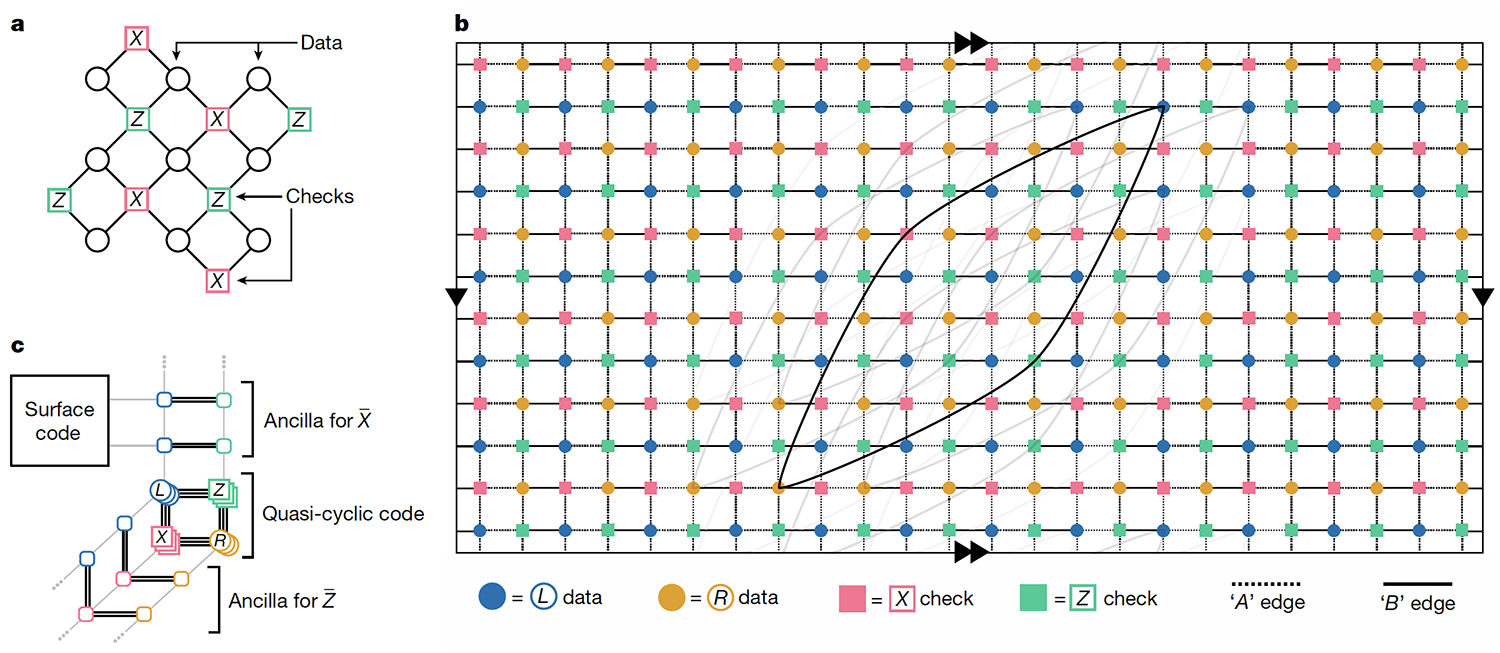
Fig. 1 | Tanner graphs of surface and BB codes.
Fig. 1 | Tanner graphs of surface and BB codes. a, Tanner graph of a surface code, for comparison. b, Tanner graph of a BB code with parameters [[144, 12, 12]] embedded into a torus. Any edge of the Tanner graph connects a data and a check vertex. Data qubits associated with the registers q(L) and q(R) are shown by blue and orange circles. Each vertex has six incident edges including four short-range edges (pointing north, south, east and west) and two long-range edges. We only show a few long-range edges to avoid clutter. Dashed and solid edges indicate two planar subgraphs spanning the Tanner graph, see the Methods. c, Sketch of a Tanner graph extension for measuring Z ˉ \={Z} and X ˉ \={X} following ref. 50, attaching to a surface code. The ancilla corresponding to the X ˉ \={X} measurement can be connected to a surface code, enabling load-store operations for all logical qubits by means of quantum teleportation and some logical unitaries. This extended Tanner graph also has an implementation in a thickness-2 architecture through the A and B edges (Methods).
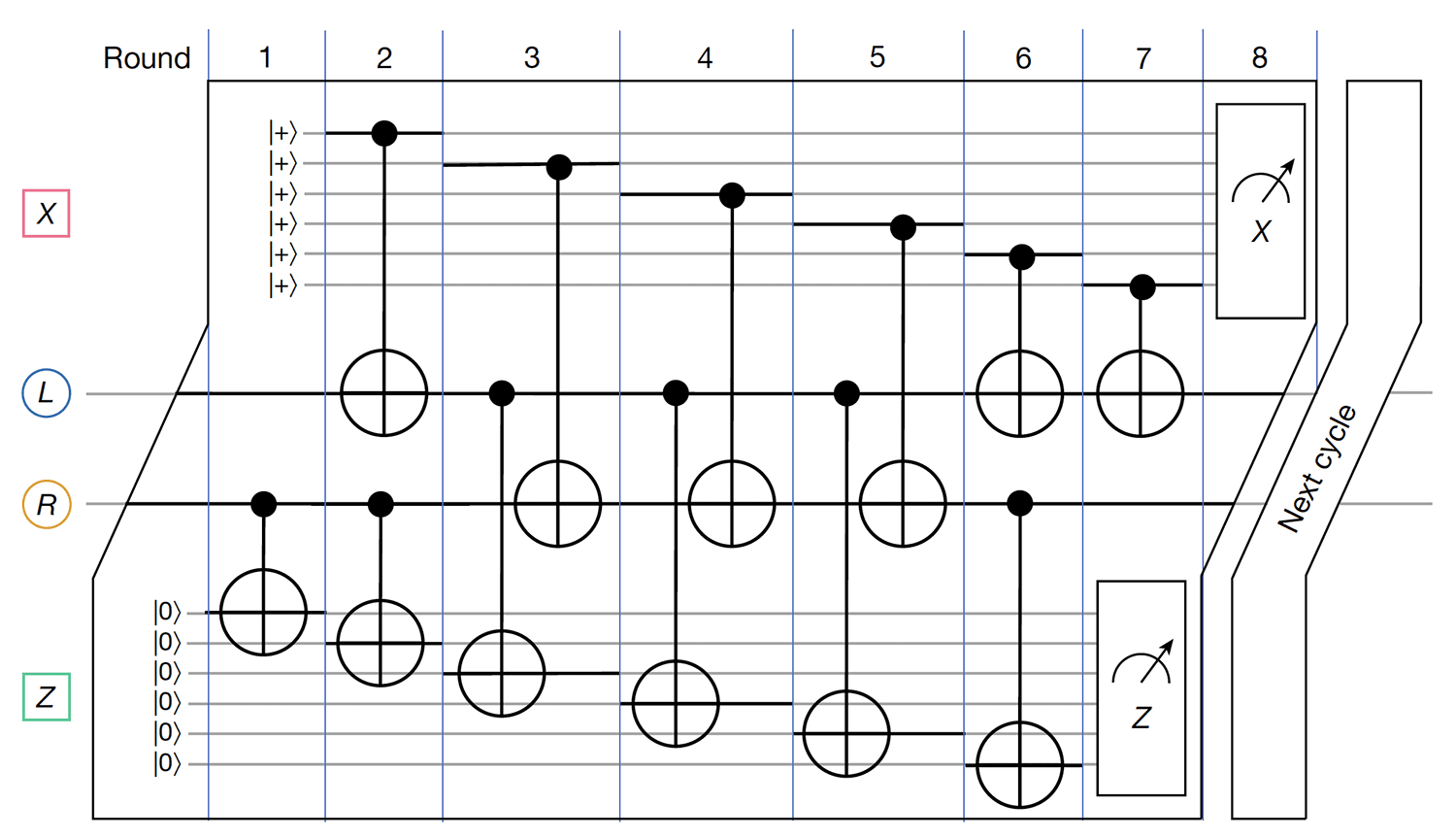
Fig. 2 | Syndrome measurement circuit.
Fig. 2 | Syndrome measurement circuit. Full cycle of syndrome measurements relying on seven layers of CNOTs. We provide a local view of the circuit that only includes one data qubit from each register q(L) and q(R) . The circuit is symmetric under horizontal and vertical shifts of the Tanner graph. Each data qubit is coupled by CNOTs with three X-check and three Z-check qubits: see the Methods for more details.
Why error correction matters
Today, our users benefit from novel error mitigation techniques — methods for reducing or eliminating the effect of noise when calculating observables, alongside our work suppressing errors at the hardware level. This work brought us into the era of quantum utility. IBM researchers and partners all over the world are exploring practical applications of quantum computing today with existing quantum systems. Error mitigation lets users begin looking for quantum advantage on real quantum hardware.
But error mitigation comes with its own overhead, requiring running the same executions repeatedly so that classical computers can use statistical methods to extract an accurate result. This limits the scale of the programs you can run, and increasing that scale requires tools beyond error mitigation — like error correction.
Last year, we debuted a new roadmap laying out our plan to continuously improve quantum computers over the next decade. This new paper is an important example of how we plan to continuously increasing the complexity (number of gates) of the quantum circuits that can be run on our hardware. It will allow us to transition from running circuits with 15,000 gates to 100 million, or even 1 billion gates.
Bravyi, S., Cross, A.W., Gambetta, J.M. et al. High-threshold and low-overhead fault-tolerant quantum memory. Nature 627, 778–782 (2024). https://doi.org/10.1038/s41586-024-07107-7
Start using our 100+ qubit systems
Keep exploring.

Exploring measurement error mitigation with the Mthree Qiskit extension
Computing with error-corrected quantum computers.

The era of quantum utility must also be the era of responsible quantum computing

Logical gates with magic state distillation

COMMENTS
We introduce GRM, a large-scale reconstructor capable of recovering a 3D asset from sparse-view images in around 0. 1s. 3D Reconstruction Image to 3D +1. 335. 0.77 stars / hour. Paper. Code. Papers With Code highlights trending Machine Learning research and the code to implement it.
This paper presents Arc2Face, an identity-conditioned face foundation model, which, given the ArcFace embedding of a person, can generate diverse photo-realistic images with an unparalleled degree of face similarity than existing models. Ranked #1 on Face Generation on AgeDB. Papers With Code highlights trending Machine Learning research and ...
By decomposing the image formation process into a sequential application of denoising autoencoders, diffusion models (DMs) achieve state-of-the-art synthesis results on image data and beyond. Ranked #2 on Layout-to-Image Generation on COCO-Stuff 256x256. Denoising Image Inpainting +5. 64,829.
Papers + Code - MIT-IBM Watson AI Lab. Research. Research. What's Next in AI. Papers + Code. Peer-review is the lifeblood of scientific validation and a guardrail against runaway hype in AI. Our commitment to publishing in the top venues reflects our grounding in what is real, reproducible, and truly innovative. Sort by Newest ↓.
Unikernels are famous for providing excellent performance in terms of boot times, throughput and memory consumption, to name a few metrics. Operating Systems. 1,933. 0.11 stars / hour. Paper. Code. Papers With Code highlights trending Computer Science research and the code to implement it.
Papers with code has 12 repositories available. Follow their code on GitHub. Papers with code has 12 repositories available. Follow their code on GitHub. Skip to content. ... Tips for releasing research code in Machine Learning (with official NeurIPS 2020 recommendations) 2,442 MIT 696 3 2 Updated May 19, 2023. galai Public
The aim of this paper is to introduce a novel two-step framework that comprises: (i) a imputation methods for statistical objects taking values in metrics spaces, and (ii) a criterion for personalizing imputation using conformal inference techniques. Methodology Applications. 0. 26 Mar 2024. Paper.
Recently papers with code and evaluation metrics. A Public and Reproducible Assessment of the Topics API on Real Data. yohhaan/topics_api_analysis • 28 Mar 2024 The central point of contention is largely around the realism of the datasets used in these analyses and their reproducibility; researchers using data collected on a small sample of users or generating synthetic datasets, while ...
💡 Collated best practices from most popular ML research repositories - now official guidelines at NeurIPS 2021! Based on analysis of more than 200 Machine Learning repositories, these recommendations facilitate reproducibility and correlate with GitHub stars - for more details, see our our blog post.. For NeurIPS 2021 code submissions it is recommended (but not mandatory) to use the README ...
Recently, Papers with Code have grown in both popularity and in terms of providing a complete ecosystem for machine learning research. You can reproduce the results by using the code, checking all the previous implementations with the model performance metrics, viewing the dataset, models, and methods used in the research paper. It is the next ...
wala/graph4code • • 21 Feb 2020. We make the toolkit to build such graphs as well as the sample extraction of the 2 billion triples graph publicly available to the community for use. 1. Paper. Code. The goal of **Code Search** is to retrieve code fragments from a large code corpus that most closely match a developer's intent, which is ...
Research code is typically created by a group of scientists and published together with academic papers to facilitate research transparency and reproducibility. For this study, we define ten ...
We introduce Codex, a GPT language model fine-tuned on publicly available code from GitHub, and study its Python code-writing capabilities. A distinct production version of Codex powers GitHub Copilot. On HumanEval, a new evaluation set we release to measure functional correctness for synthesizing programs from docstrings, our model solves 28.8% of the problems, while GPT-3 solves 0% and GPT-J ...
Linked Papers With Code. In this paper, we introduce (LPWC), an RDF knowledge graph that provides comprehensive, current information about almost 400,000 machine learning publications. This includes the tasks addressed, the datasets utilized, the methods implemented, and the evaluations conducted, Papers With Code.
Download a PDF of the paper titled Code Llama: Open Foundation Models for Code, by Baptiste Rozi\`ere and 25 other authors. ... We release Code Llama under a permissive license that allows for both research and commercial use. Subjects: Computation and Language (cs.CL) Cite as: arXiv:2308.12950 [cs.CL]
Authors can sync their code to display on arXiv abstract pages from either their arXiv user account page or the Papers with Code interface, ... This expansion was driven by the broader research community's demand. After the October release, researchers beyond ML expressed a desire for easy access to relevant code. Researchers in physics ...
These are the steps that we should follow while implementing the code: →Load the the data set containing real images. →Create a random two dimensional tensor (probability distribution of fake data). →Create a discriminator model and generator model. →Train the discriminator on real and fake images.
Step 1 - Initial coding. The first step of the coding process is to identify the essence of the text and code it accordingly. While there are various qualitative analysis software packages available, you can just as easily code textual data using Microsoft Word's "comments" feature.
Usually, older papers describe simpler concepts, which is a big plus for you as a beginner. These weeks I have been reading papers on Super-Resolution, so I've chosen this 2016-year research to reproduce it with you — Accelerating the Super-Resolution Convolutional Neural Network. It describes one of the first CNN-based models for a Super ...
The code for training the machine learning models, analyzing the models, and generating the figures has been deposited to Zenodo under accession code 10653704 93. References Tieman, D. et al.
ReALM: Reference Resolution As Language Modeling. Joel Ruben Antony Moniz, Soundarya Krishnan, Melis Ozyildirim, Prathamesh Saraf, Halim Cagri Ates, Yuan Zhang, Hong Yu, Nidhi Rajshree. Reference resolution is an important problem, one that is essential to understand and successfully handle context of different kinds. This context includes both ...
Moreover, due to the limited amount of research on the high-frequency quasi-periodic oscillations (HFQPOs) phenomenon and its generation mechanisms around particles near wormholes using observational data, this paper aims to study theoretical models that can simultaneously describe both BH and wormholes by fitting observational data.
IBM has created a quantum error-correcting code about 10 times more efficient than prior methods. ... In our new paper, we introduce a new code, ... And they are going to shape not only our research going forward, but how we architect physical quantum systems. The gross code.