
- 2022 Best Tech
- Creating a Lasting Impact on the Future of Sight
- Opt-Out Screening for STIs
- RSV Roundtable
- Food Insecurity and the Dangers of Infant Formula Dilution
- Opt-Out Chlamydia Screening in Adolescent Care
- Pediatric ADHD: Zeroing in on an Untenable Burden
- The Role of the Healthcare Provider Community in Increasing Public Awareness of RSV in All Infants
- Update in Pediatric COVID-19 Vaccines

- Publications
- Conferences
- Partnerships

Case Studies
If you are interested in submitting a Puzzler of your own, please see Puzzler guidelines here .

Quiz: Can you diagnose these muscle spasms in a 19-year-old-male?
Take this Contemporary Pediatrics quiz, and see if you can correctly diagnose the patient in this case study. Submit your answer to see if you were correct.

11-year-old boy with testicular pain and rash
An 11-year-old boy presented to the emergency department complaining of left testicular pain for 2 days, described as intermittent and stabbing, which ranged between 5 and 8 of 10 in intensity. Read the full case to see if you can correctly diagnose the patient.
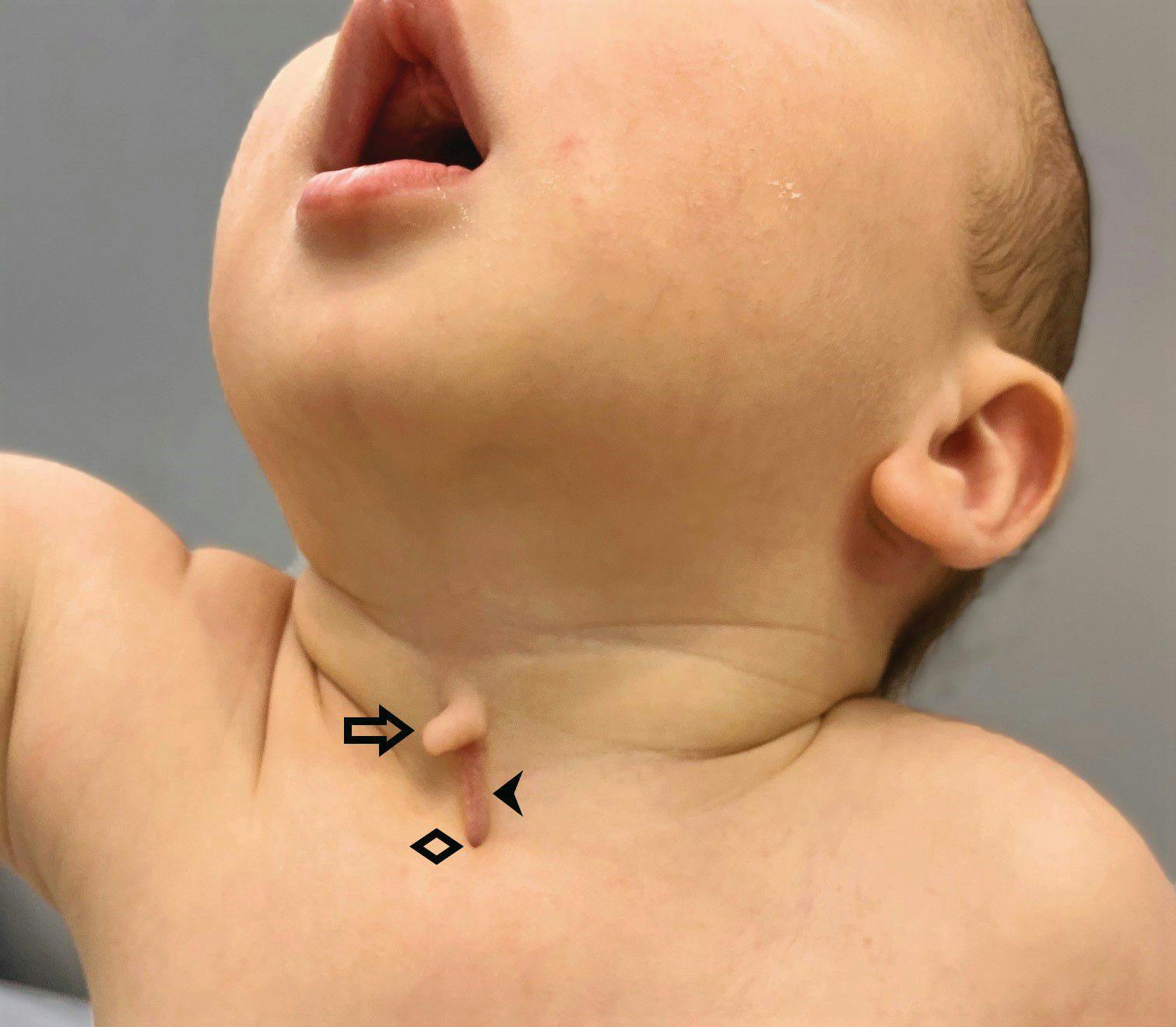
Newborn with midline neck lesion
A 4-day-old boy with a midline neck lesion was born at term by normal vaginal delivery. After birth, mid line lesion had the configuration of a linear cleft with a cephalocaudal orientation, extending from the level below the hyoid bone to the suprasternal notch with a length of 2.5 cm and a width of 0.5 cm. What's the diagnosis?
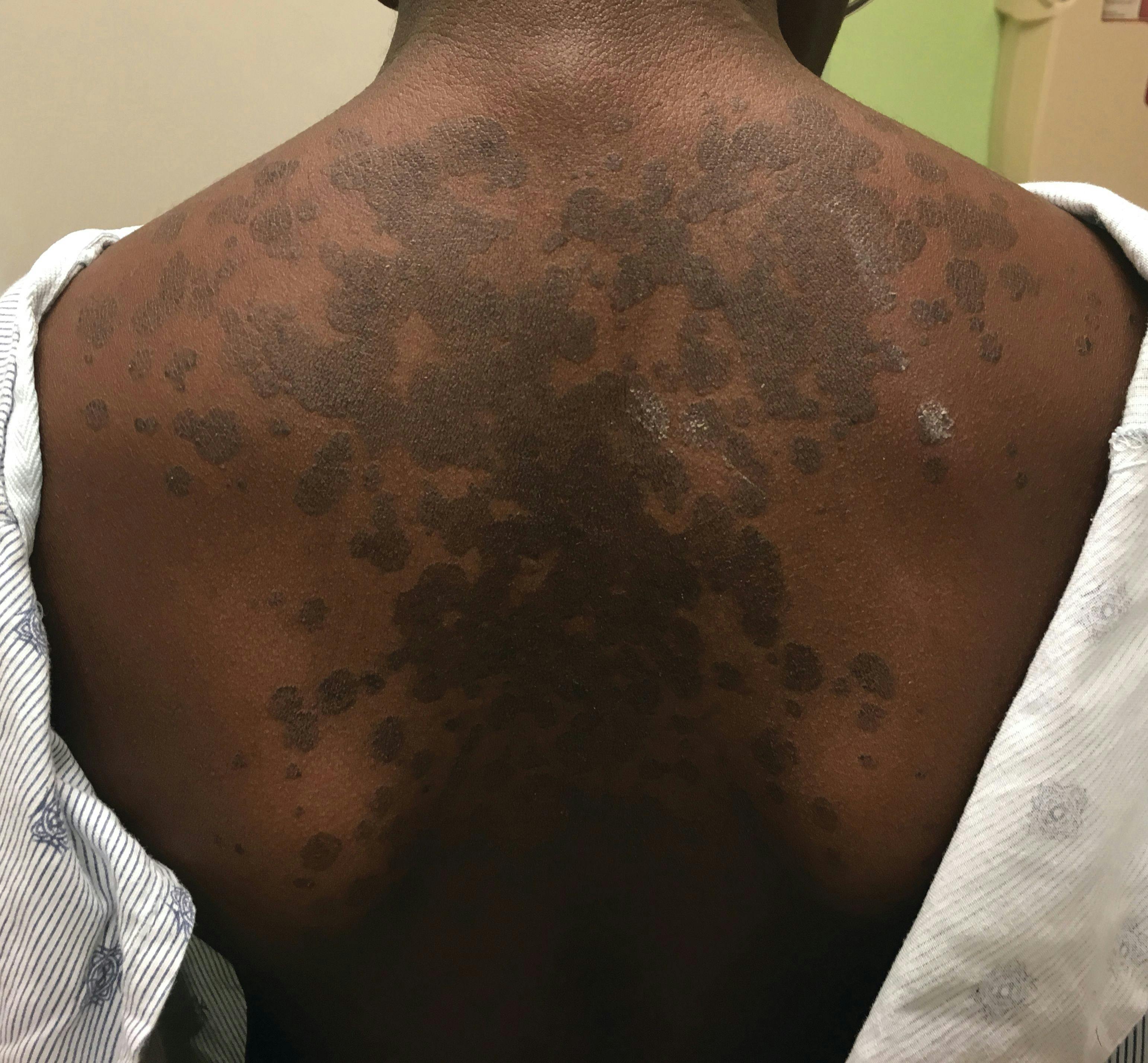
A 13-year-old girl with well-demarcated rash on back and chest
A healthy 13-year-old girl presented with a 1-month history of an asymptomatic, well-demarcated rash on her back and upper chest. The eruption consisted of discrete, dark brown papules that coalesced into large, flat-topped plaques with mild superficial scale and accentuation of skin markings. What's the diagnosis?

Suspicious facial swelling in a 22-month-old girl
A 22-month-old female patient with sickle cell disease on folic acid and penicillin prophylaxis with a 3-day history of nasal congestion, rhinorrhea, fever and decreased oral intake presents to the emergency department (ED) for acute facial swelling noted when she woke up from a nap. What's the diagnosis?
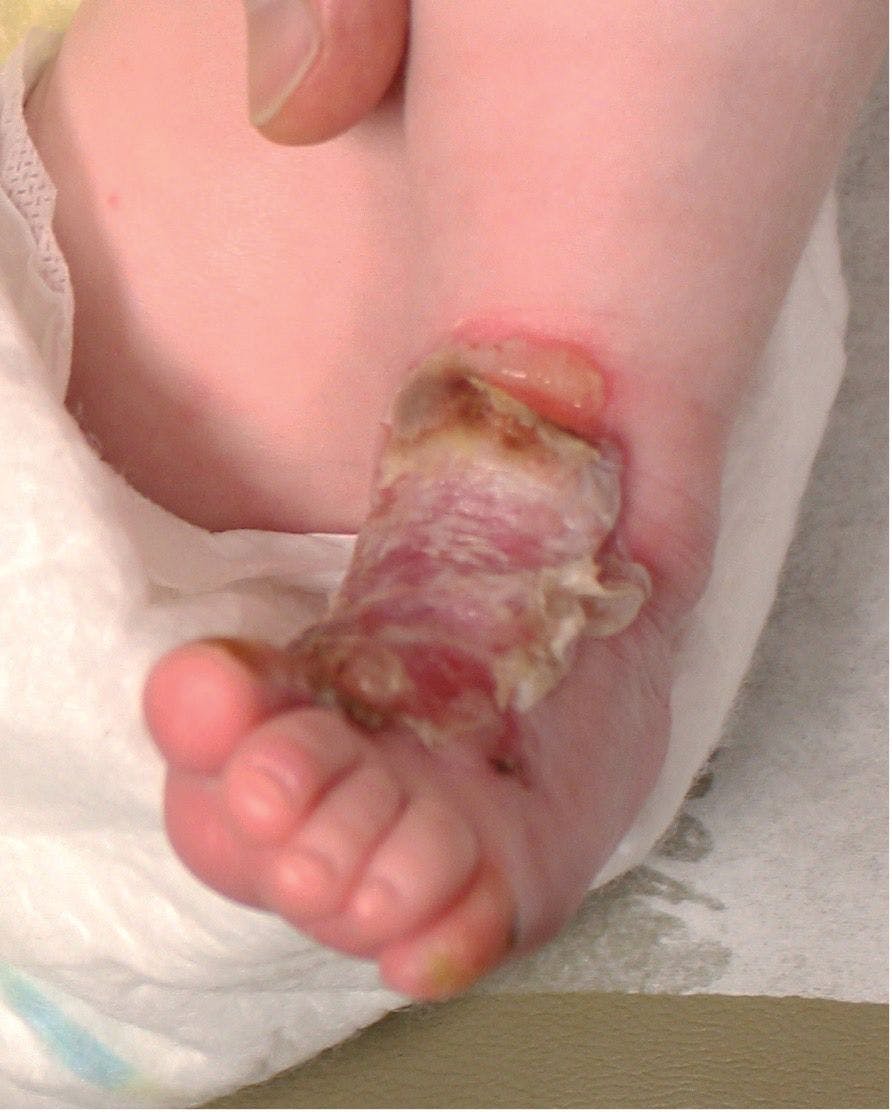
Friction-induced blistering on a child’s feet
You are called to the hospital nursery to evaluate a healthy full-term newborn boy who developed painful flaccid blisters and erosions on the tops of his feet and ankles shortly after birth. His mother had a history of similar recurrent skin lesions that healed with scarring. She also had oral and gastrointestinal tract involvement. What's the diagnosis?

Hypothermia and abnormal eye movements in a 5-week-old infant
A 5-week-old female infant born at 38 weeks presents to her pediatrician with abnormal eye movements. What’s the diagnosis?

Neonate experiences coffee ground emesis
The infant did not show signs of illness; her mother experienced a routine pregnancy and prenatal lab test results were normal. What is the diagnosis?
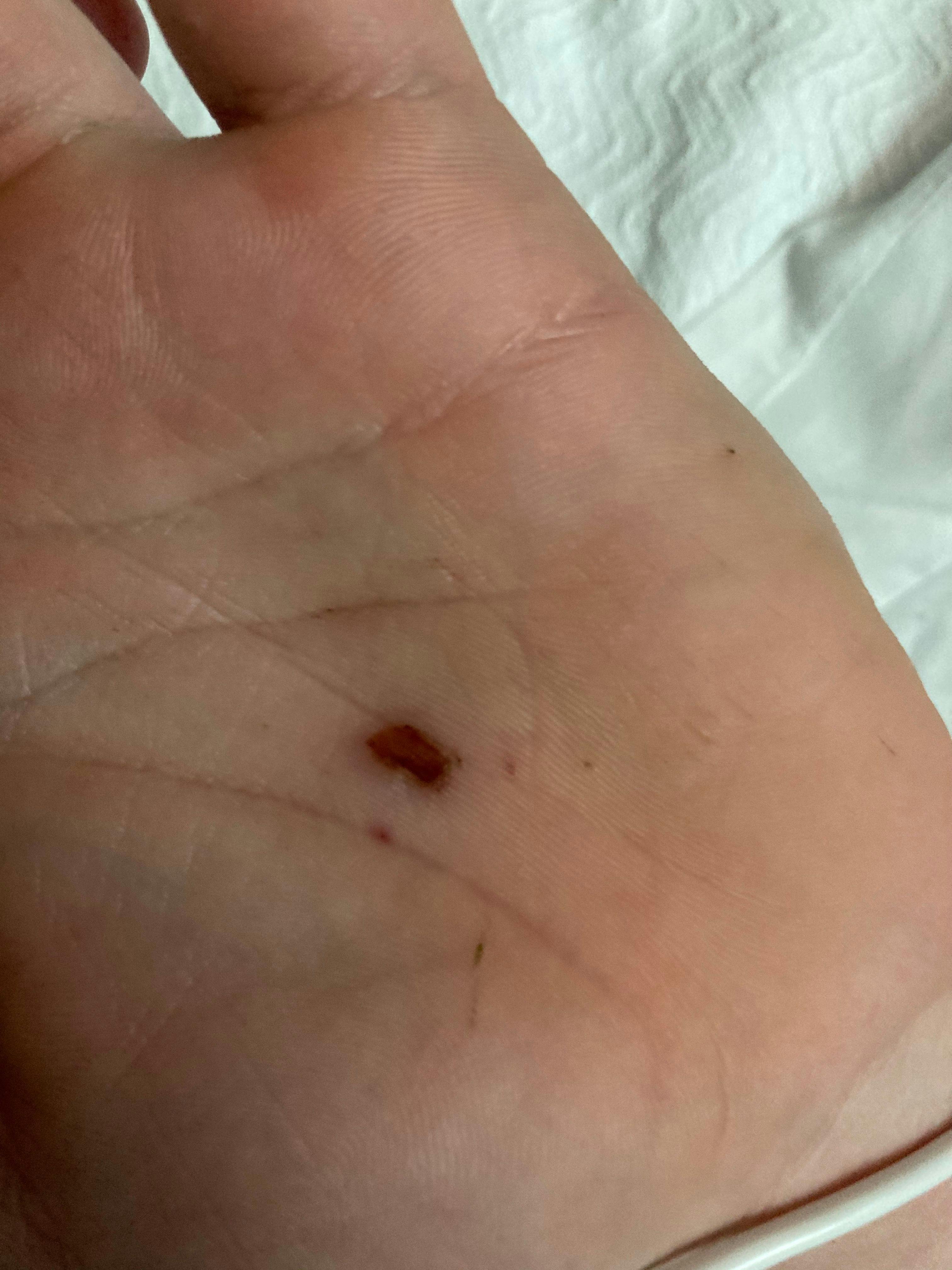
Muscle spasms in a 19-year-old male
A 19-year-old male presents to the emergency department (ED) with headache and fever of 4 days’ duration. Six days earlier, his left palm had been punctured by a rusty nail. What's the diagnosis?

Treating an 18-month-old who tested positive for cannabis exposure
As more and more states legalize recreational marijuana, caregivers need to be vigilant about keeping products out of reach of children.

Scalp thickening and folding in a pubertal boy
A 16-year-old boy with developmental delay and intellectual disability developed dramatic chronic wrinkling of his scalp over a year ago. The lesions were persistent but not symptomatic. What's the diagnosis?
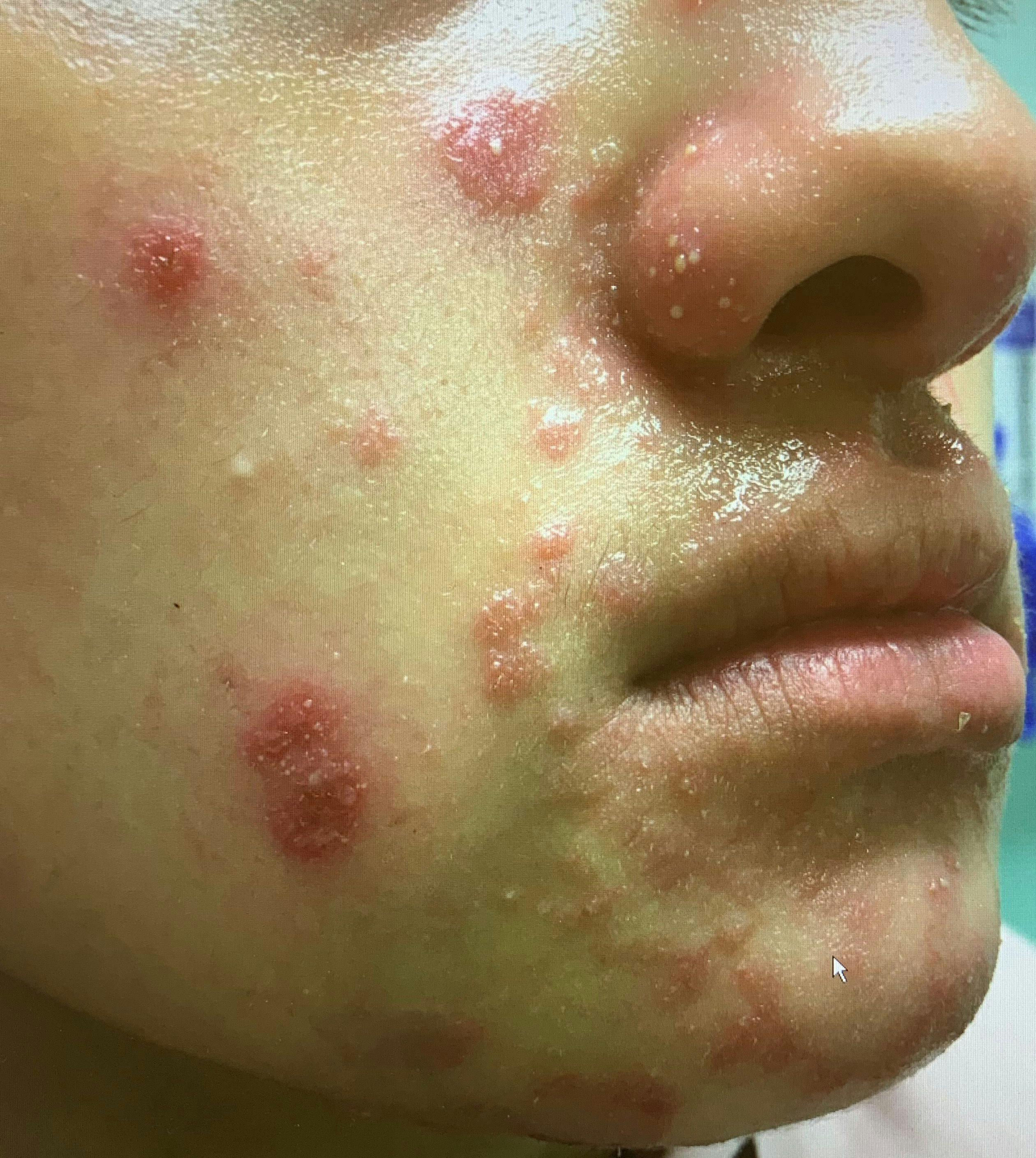
Worsening acne after isotretinoin treatment in an adolescent girl
A healthy 17-year-old girl with inflammatory acne had failed to respond to topical tretinoin, benzoyl peroxide, and oral minocycline. What's the diagnosis?

Psychosis in an 18-year-old male
Alex, an 18-year-old male, presented to the emergency department with a 4-day history of paranoia, agitation, and disorganized behavior. He had no psychiatric history or prior mental health contact and no known medical conditions.
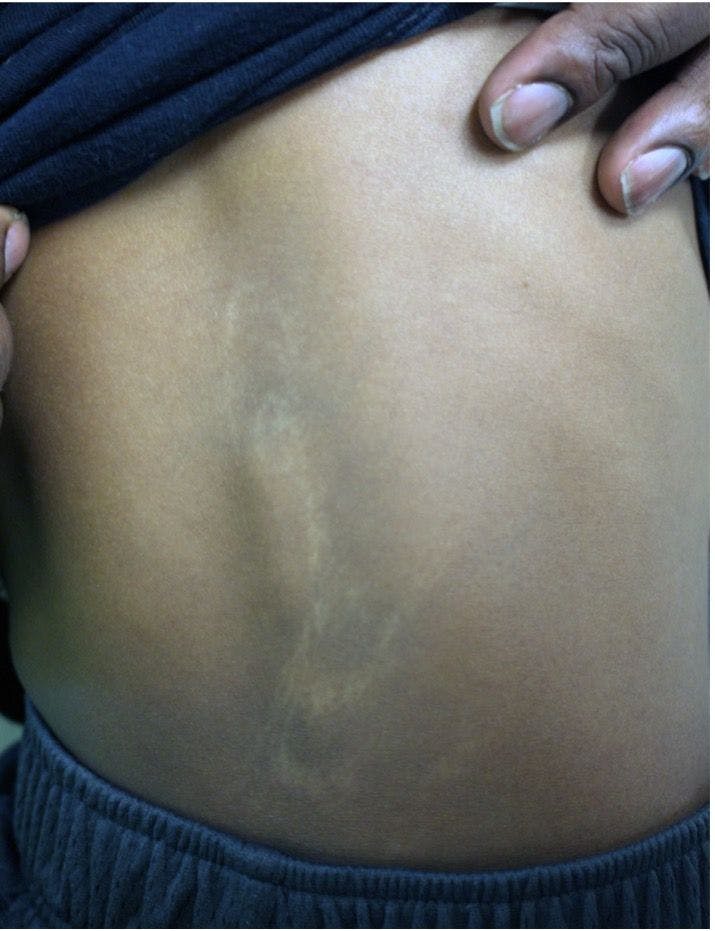
Congenital hypopigmented macules on a healthy child
You are asked to evaluate an African American boy aged 4 years with a birthmark on his back and right arm. He is healthy with normal growth and development. What's the diagnosis?
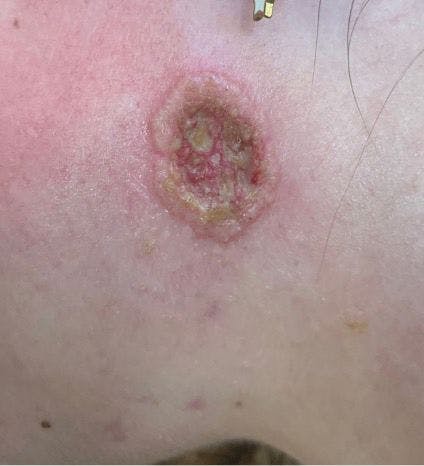
Just a little birthmark?
You are asked to evaluate a healthy 1.5-day-old girl who has a congenital red patch with coarse telangiectasias and a surrounding ring of pallor on the right shoulder. What's your diagnosis?

Can intranasal corticosteroids improve obstructive sleep apnea syndrome in children?
A study featured at CHEST 2022 investigated INCS in children with OSAS to determine if the therapy improved their symptoms, polysomnography findings, behavior, and quality of life.
Case of inflammatory acne or something else?
This case study analyzes variants of majocchi granuloma and potential treatment.

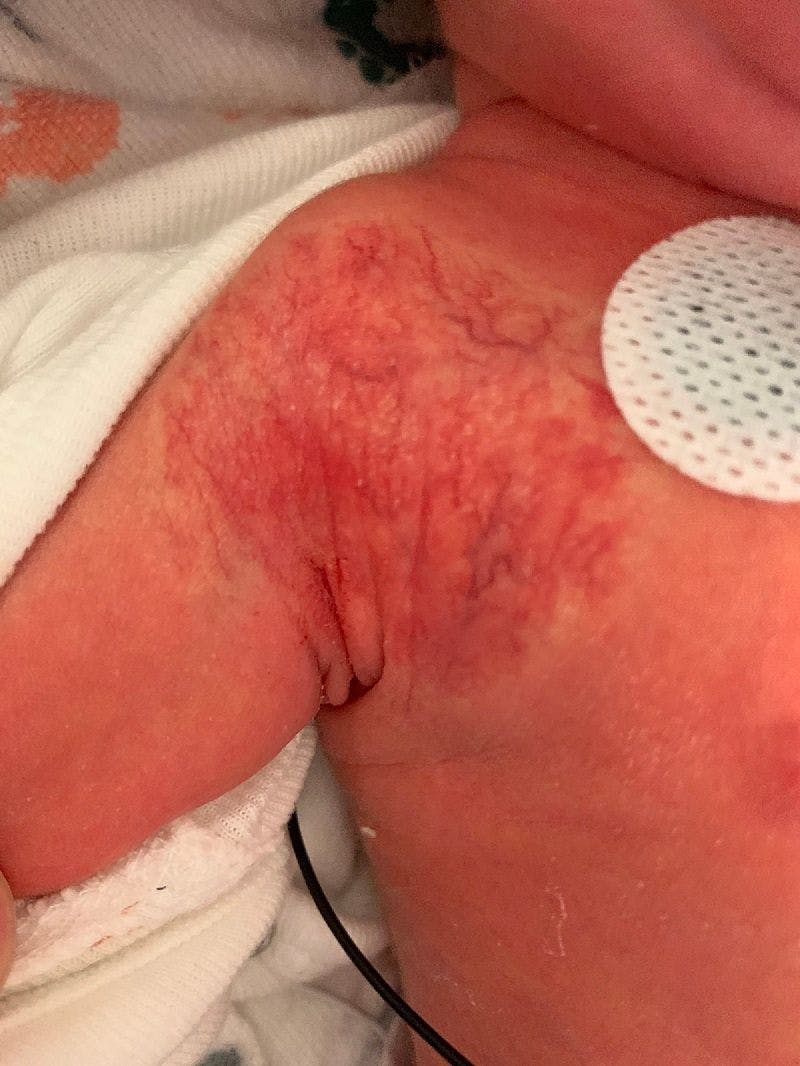
A case of late-onset group B Streptococcus infection in fraternal twins
A 29-year-old White woman presented to the labor and delivery unit due to preterm premature rupture of membranes and delivered twins. The twins were transferred to the neonatal intensive care unit following delivery.
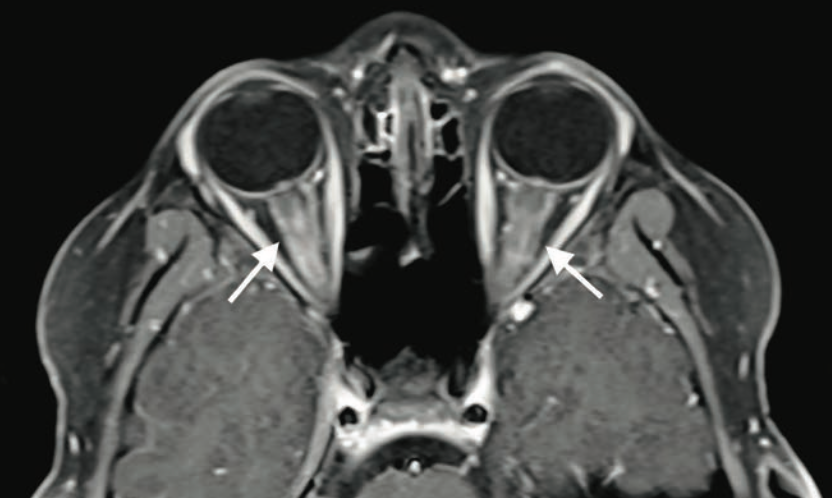
Bilateral blurry vision in a 9-year-old boy
A 9-year-old boy with no significant medical history presented to the emergency department with 2 days of painless blurry vision. What's the diagnosis?
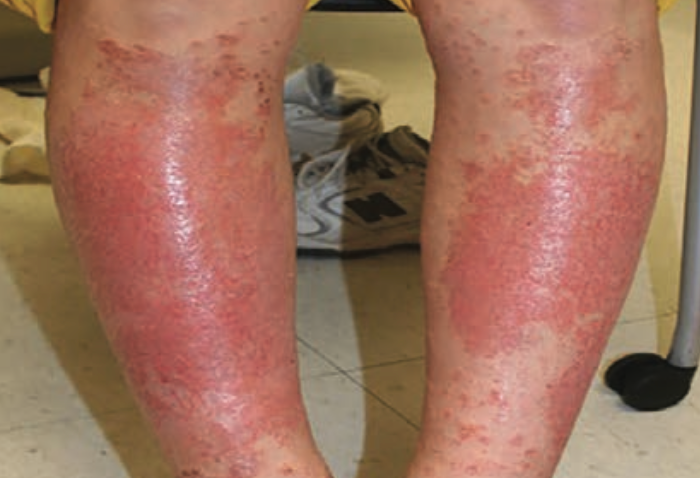
A case of shin guard dermatitis?
A healthy 14-year old boy was evaluated for an intensely pruritic shin rash that developed 2 weeks prior and had been treated with oral antibiotics for 10 days.

A renal anomaly? Look for a Müllerian anomaly as well
A retrospective study found that postmenarchal women with a renal anomaly were also at risk of having a Müllerian anomaly.

A case of progressive joint pain and rash in a 5-year-old
A 5-year-old nonverbal boy with autism spectrum disorder and global developmental delay presented to the emergency department with bilateral lower-extremity bruising and progressive difficulty ambulating. What's the diagnosis?
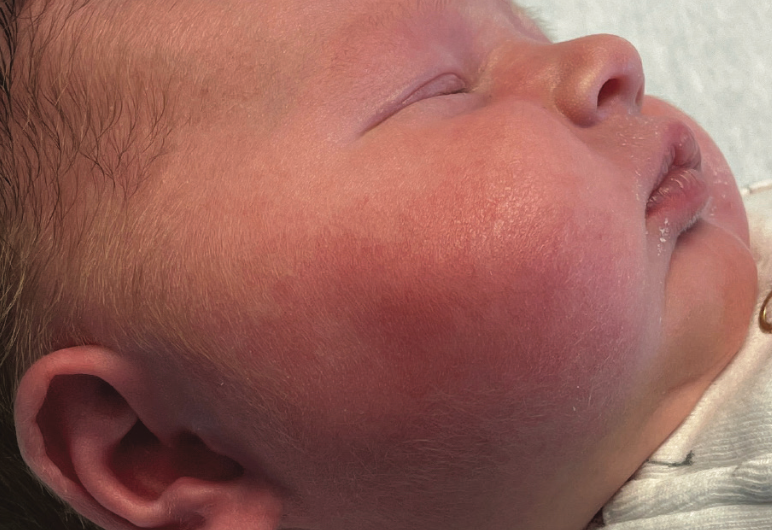
Fever and facial swelling in a neonate
An 18-day-old girl whose right cheek had become increasingly red and warm over 24 hours was directly admitted to an inpatient unit. She had firmness and pain to the affected area, fussiness, increased sleeping, and poor feeding, preferring the bottle to breastfeeding. What's the diagnosis?

Altered mental state in a 2-year-old boy
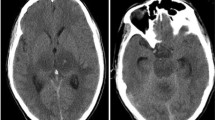
A 26-month-old boy presents for mild altered mental status and balance issues following a fall the day before. There was no loss of consciousness or vomiting but he subsequently complained of left-sided head pain. What's the diagnosis?
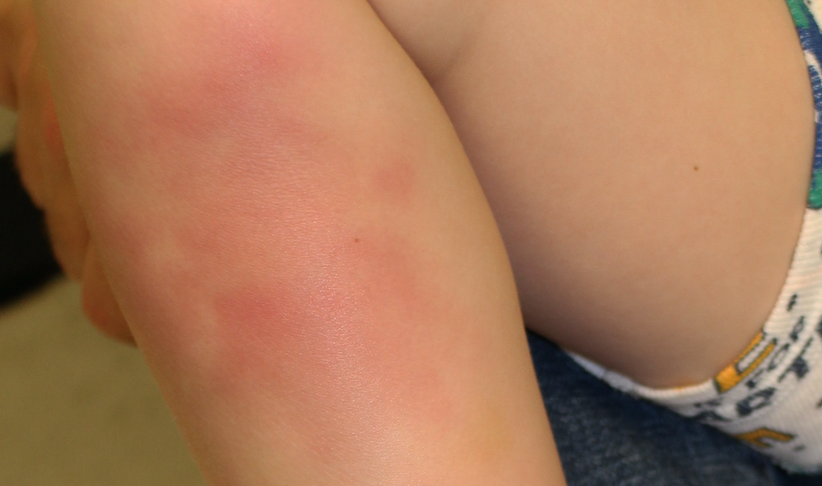
Painful red lumpy leg
A healthy 4-year-old boy presents with painful deep-seated bumps on the front of his right leg. He also complains of a sore throat for the last 4 days. What's the diagnosis?
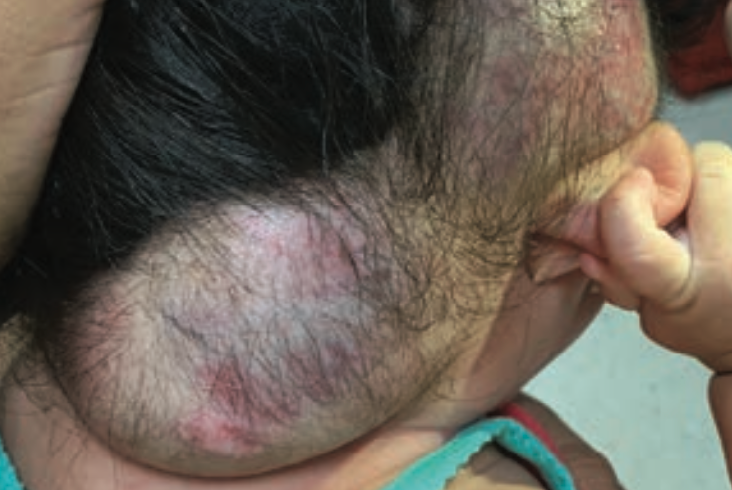
Severe hemorrhage from infantile hemangioma
A 5-month-old girl with a large scalp infantile hemangioma (IH), present since 6 weeks of age, is evaluated in the emergency department for lethargy and pallor.
Generalized, eruptive lichen planus in a pediatric patient
A healthy 14-year-old boy presented at our dermatology practice with acute onset of an intensely itchy rash that first appeared 2 months prior.
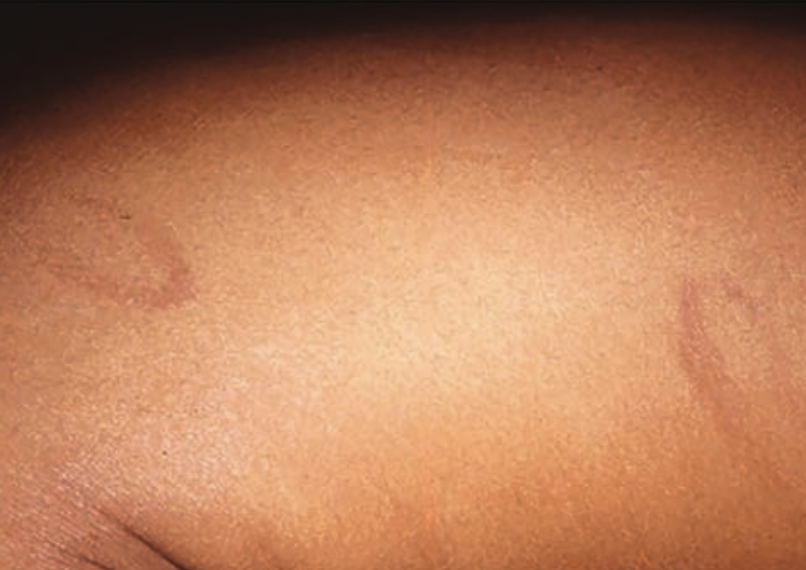
Annular scars with hyperpigmentation
An 18-month-old girl presents with ringed scars with hyperpigmentation on the right side of her buttocks and back. What's the diagnosis?
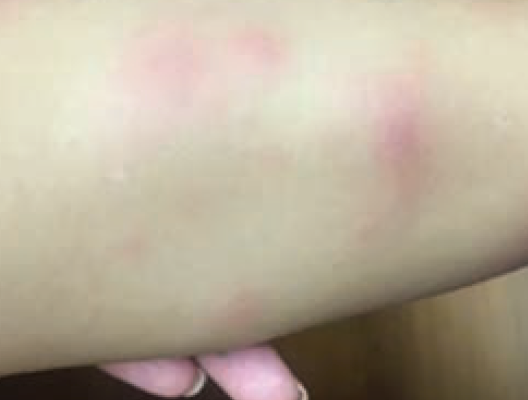
Lower-extremity nodules in a 2-year-old girl
A 2-year-old girl presents with an itchy, bilateral leg rash. Additionally, the child had several bruises that felt like "hard welts" and were warm to the touch. What's the diagnosis?
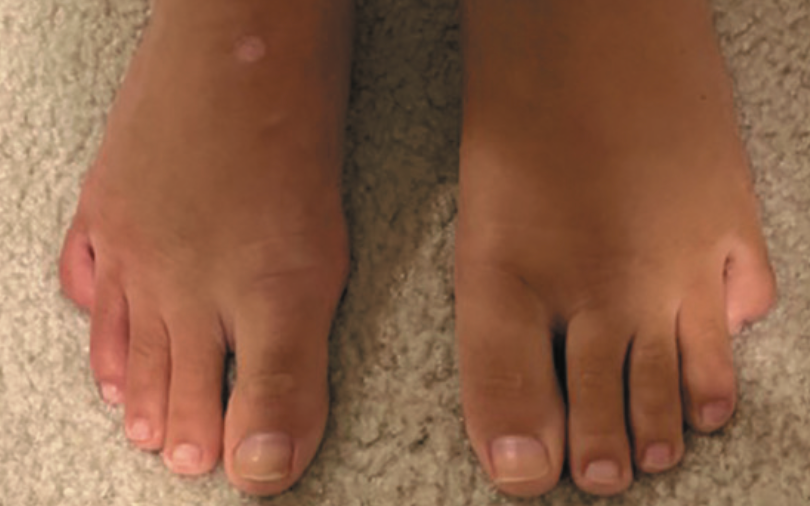
Persistent foot and leg swelling in a 17-year-old female
A 17-year-old girl presents with a 2-year history of unilateral swelling of the left lower extremity as well as a poorly healed ankle sprain of the affected extremity 3 years prior that slowly resolved but left persistent swelling. What's the diagnosis?
2 Commerce Drive Cranbury, NJ 08512
609-716-7777

Paediatrics
Showing results 1 - 10 of 100
Sorted by most recent
16 April 2024
11 April 2024
8 April 2024
4 April 2024
2 April 2024
21 March 2024
19 March 2024
18 March 2024
We have a new app!
Take the Access library with you wherever you go—easy access to books, videos, images, podcasts, personalized features, and more.
Download the Access App here: iOS and Android . Learn more here!
- Remote Access
- Save figures into PowerPoint
- Download tables as PDFs
- Case Files®: Pediatrics
- Pediatrics Examination and Board Review
- Neonatal-Perinatal Medicine: Specialty Board Review
- Symptom-Based Diagnosis in Pediatrics

Case Files: Pediatrics, 6e
Author(s): Eugene C. Toy; Mark D. Hormann; Robert J. Yetman; Margaret C. McNeese; Sheela L. Lahoti; Emma A. Omoruyi; Abby M. Geltemeyer
- 2 Infant of a Diabetic Mother
- 3 Neonatal Hyperbilirubinemia
- 4 Sepsis and Group B Streptococcal Infections
- 6 Neonatal Herpes Simplex Virus Infection
- 8 Transient Tachypnea of the Newborn
- 10 Failure to Thrive
- 38 Child Abuse
- 14 Pneumonia
- 17 Pelvic Inflammatory Disease
- 27 Bacterial Meningitis
- 28 Bacterial Enteritis
- 40 Immunodeficiency
- 54 Appendicitis
- 55 Acute Epstein-Barr Virus (Infectious Mononucleosis)
- 29 Subdural Hematoma
- 30 Complex Febrile Seizure
- 48 Migraine Without Aura
- 56 Myasthenia Gravis
- 60 Concussion
- 22 Patent Ductus Arteriosus With Heart Failure
- 23 Truncus Arteriosus
- 18 Cystic Fibrosis
- 20 Asthma Exacerbation
- 61 Electronic Cigarette or Vaping Induced Acute Lung Injury (EVALI) Online Only
- 7 Esophageal Atresia
- 15 Rectal Bleeding
- 34 Intestinal Malrotation
- 53 Inflammatory Bowel Disease
- 35 Posterior Urethral Valves
- 52 Acute Postinfectious Glomerulonephritis
- 11 Anemia in the Pediatric Patient
- 13 Sickle Cell Disease With Vaso-Occlusive Crisis
- 19 Acute Lymphoblastic Leukemia
- 33 Suspected Neuroblastoma
- 42 Diabetic Ketoacidosis
- 44 Growth Hormone Deficiency
- 45 Precocious Puberty
- 41 Trisomy 21
- 50 Turner Syndrome
- 37 Immune Thrombocytopenic Purpura
- 39 Kawasaki Disease
- 51 Systemic Lupus Erythematosus
- 57 Oligoarticular Juvenile Idiopathic Arthritis
- 58 Anaphylactoid Purpura (Henoch-Schönlein Purpura)
- 49 Adolescent Substance Use Disorder
- 62 Alopecia Areata Online Only
- 5 Accidental Ingestion of Opioids
- 25 Lead Ingestion (Microcytic Anemia)
- 59 Attention Deficit Hyperactivity Disorder
- 31 Duchenne Muscular Dystrophy
- 36 Nursemaid’s Elbow (Radial Head Subluxation)
- 47 Slipped Capital Femoral Epiphysis
- 1 Infant Rashes
- 26 Stevens-Johnson Syndrome
- 32 Atopic Dermatitis
- 9 Congenital Cataracts
- 16 Acute Otitis Media
- 43 Obstructive Sleep Apnea Syndrome
- 46 Retropharyngeal Abscess
- 21 Sudden Infant Death Syndrome
- Accidental Ingestion of Opioids
- Acute Epstein-Barr Virus (Infectious Mononucleosis)
- Acute Lymphoblastic Leukemia
- Acute Otitis Media
- Acute Postinfectious Glomerulonephritis
- Adolescent Substance Use Disorder
- Alopecia Areata Online Only
- Anaphylactoid Purpura (Henoch-Schönlein Purpura)
- Anemia in the Pediatric Patient
- Appendicitis
- Asthma Exacerbation
- Atopic Dermatitis
- Attention Deficit Hyperactivity Disorder
- Bacterial Enteritis
- Bacterial Meningitis
- Child Abuse
- Complex Febrile Seizure
- Congenital Cataracts
- Cystic Fibrosis
- Diabetic Ketoacidosis
- Duchenne Muscular Dystrophy
- Electronic Cigarette or Vaping Induced Acute Lung Injury (EVALI) Online Only
- Esophageal Atresia
- Failure to Thrive
- Growth Hormone Deficiency
- Immune Thrombocytopenic Purpura
- Immunodeficiency
- Infant of a Diabetic Mother
- Infant Rashes
- Inflammatory Bowel Disease
- Intestinal Malrotation
- Kawasaki Disease
- Lead Ingestion (Microcytic Anemia)
- Migraine Without Aura
- Myasthenia Gravis
- Neonatal Herpes Simplex Virus Infection
- Neonatal Hyperbilirubinemia
- Nursemaid’s Elbow (Radial Head Subluxation)
- Obstructive Sleep Apnea Syndrome
- Oligoarticular Juvenile Idiopathic Arthritis
- Patent Ductus Arteriosus With Heart Failure
- Pelvic Inflammatory Disease
- Posterior Urethral Valves
- Precocious Puberty
- Rectal Bleeding
- Retropharyngeal Abscess
- Sepsis and Group B Streptococcal Infections
- Sickle Cell Disease With Vaso-Occlusive Crisis
- Slipped Capital Femoral Epiphysis
- Stevens-Johnson Syndrome
- Subdural Hematoma
- Sudden Infant Death Syndrome
- Suspected Neuroblastoma
- Systemic Lupus Erythematosus
- Transient Tachypnea of the Newborn
- Truncus Arteriosus
- Turner Syndrome
- Case report
- Open access
- Published: 21 February 2018
Pediatric severe asthma: a case series report and perspectives on anti-IgE treatment
- Virginia Mirra 1 ,
- Silvia Montella 1 &
- Francesca Santamaria 1
BMC Pediatrics volume 18 , Article number: 73 ( 2018 ) Cite this article
11k Accesses
11 Citations
12 Altmetric
Metrics details
The primary goal of asthma management is to achieve disease control for reducing the risk of future exacerbations and progressive loss of lung function. Asthma not responding to treatment may result in significant morbidity. In many children with uncontrolled symptoms, the diagnosis of asthma may be wrong or adherence to treatment may be poor. It is then crucial to distinguish these cases from the truly “severe therapy-resistant” asthmatics by a proper filtering process. Herein we report on four cases diagnosed as difficult asthma, detail the workup that resulted in the ultimate diagnosis, and provide the process that led to the prescription of omalizumab.
Case presentation
All children had been initially referred because of asthma not responding to long-term treatment with high-dose inhaled steroids, long-acting β 2 -agonists and leukotriene receptor antagonists. Definitive diagnosis was severe asthma. Three out four patients were treated with omalizumab, which improved asthma control and patients’ quality of life. We reviewed the current literature on the diagnostic approach to the disease and on the comorbidities associated with difficult asthma and presented the perspectives on omalizumab treatment in children and adolescents. Based on the evidence from the literature review, we also proposed an algorithm for the diagnosis of pediatric difficult-to-treat and severe asthma.
Conclusions
The management of asthma is becoming much more patient-specific, as more and more is learned about the biology behind the development and progression of asthma. The addition of omalizumab, the first targeted biological treatment approved for asthma, has led to renewed optimism in the management of children and adolescents with atopic severe asthma.
Peer Review reports
Children with poor asthma control have an increased risk of severe exacerbations and progressive loss of lung function, which results in the relevant use of health resources and impaired quality of life (QoL) [ 1 ]. Therefore, the primary goal of asthma management at all ages is to achieve disease control [ 2 , 3 , 4 ].
According to recent international guidelines, patients with uncontrolled asthma require a prolonged maintenance treatment with high-dose inhaled corticosteroids (ICS) in association with a long-acting β 2 -agonist (LABA) plus oral leukotriene receptor antagonist (LTRA) (Table 1 ) [ 5 ].
Nevertheless, in the presence of persistent lack of control, reversible factors such as adherence to treatment or inhalation technique should be first checked for, and diseases that can masquerade as asthma should be promptly excluded. Finally, additional strategies, in particular anti-immunoglobulin E (anti-IgE) treatment (omalizumab), are suggested for patients with moderate or severe allergic asthma that remains uncontrolled in Step 4 [ 5 ].
Herein, we reviewed the demographics, clinical presentation and treatment of four patients with uncontrolled severe asthma from our institution in order to explain why we decided to prescribe omalizumab. We also provided a review of the current literature that focuses on recent advances in the diagnosis of pediatric difficult asthma and the associated comorbidities, and summarizes the perspectives on anti-IgE treatment in children and adolescents.
Case presentations
Table 2 summarizes the clinical characteristics and the triggers/comorbidities of the cases at referral to our Institution. Unfortunately, data on psychological factors, sleep apnea, and hyperventilation syndrome were not available in any case. Clinical, lung function and airway inflammation findings at baseline and after 12 months of follow-up are reported in Table 3 . In the description of our cases, we used the terminology recommended by the ERS/ATS guidelines on severe asthma [ 6 ].
A full-term male had severe preschool wheezing and, since age 3, recurrent, severe asthma exacerbations with frequent hospital admissions. At age 11, severe asthma was diagnosed. Sensitization to multiple inhalant allergens (i.e., house dust mites, dog dander, Graminaceae pollen mix, and Parietaria judaica ) and high serum IgE levels (1548 KU/l) were found. Body mass index (BMI) was within normal range. Combined treatment with increasing doses of ICS (fluticasone, up to 1000 μg/day) in association with LABA (salmeterol, 100 μg/day) plus LTRA (montelukast, 5 mg/day) has been administered over 2 years. Nevertheless, persistent symptoms and monthly hospital admissions due to asthma exacerbations despite correct inhaler technique and good adherence were reported. Parents refused to perform any test to exclude gastroesophageal reflux (GER) as comorbidity [ 6 ]. However, an ex-juvantibus 2-month-course with omeprazole was added to asthma treatment [ 7 ], but poor control persisted. Anterior rhinoscopy revealed rhinosinusitis that was treated with nasal steroids for six months [ 8 ], but asthma symptoms were unmodified. Treatment with omalizumab was added at age 12. Reduced hospital admissions for asthma exacerbations, no further need for systemic steroids, and improved QoL score (from 2.0 up to 6.7 out of a maximum of 7 points) were documented over the following months. Unfortunately, after one year of treatment, adherence to omalizumab decreased because of family complaints, and eventually parents withdrew their informed consent and discontinued omalizumab. Currently, by age 17, treatment includes inhaled salmeterol/fluticasone (100 μg/500 μg∙day -1 , respectively) plus oral montelukast (10 mg/day). Satisfactory symptom control is reported, with no asthma exacerbations.
A full-term male, who had a recurrent severe preschool wheezing, at 6 years of age developed exercise-induced asthma. At age 10, severe asthma was diagnosed. High serum IgE levels (1300 KU/l) and skin prick tests positive to house dust mites were found. Despite a 3-year treatment with progressively increasing doses of inhaled fluticasone (up to 1000 μg/day) combined with salmeterol (100 μg/day) and oral montelukast (5 mg/day), monthly hospital admissions with systemic steroids use were reported. At age 13, a 24-h esophageal impedance/pH study demonstrated the presence of acid and non-acid GER [ 7 ]. Esomeprazole was added to asthma medications, but with an incomplete clinical benefit for respiratory symptoms. Esomeprazole was withdrawn after 3 months, and parents refused to re-test for GER. As respiratory symptoms persisted uncontrolled despite treatment, severe asthma was definitively diagnosed [ 6 ]. BMI was within the normal range and anterior rhinoscopy excluded rhinosinusitis. Inhaler technique and adherence were good; thus we considered the anti-IgE treatment option [ 9 ]. Subcutaneous omalizumab was started, with fast improvement of both symptoms and QoL score (from 3.9 up to 6.5). Seventeen months later, the dose of ICS had been gradually tapered and oral montelukast definitely discontinued. Currently, at age 14, treatment includes the combined administration of bimonthly subcutaneous omalizumab and of daily inhaled salmeterol/fluticasone (50 μg/100 μg∙day - 1 , respectively). Asthma control is satisfactory and no side effects are reported. Omalizumab has been continuously administered for 2.6 years and is still ongoing.
A full-term male had severe preschool wheezing and, since age 3, recurrent, severe asthma exacerbations with acute respiratory failure that frequently required intensive care unit (ICU) admission. At age 6, sensitization to multiple perennial inhalant (i.e., house dust mites, dog and cat danders, Alternaria alternata , Graminaceae pollen mix, Artemisia vulgaris , Parietaria judaica , and Olea europaea pollen) and food allergens (i.e., egg, milk, and peanut) was diagnosed. Serum IgE levels were 2219 KU/l. Weight and height were appropriate for age and sex. The patient has been treated over 3 years with a combined scheme of high-dose inhaled fluticasone (up to 1000 μg/day) plus salmeterol (100 μg/day) and oral montelukast (5 mg/day), with correct inhaler technique and good adherence. Despite this, monthly hospital admissions with systemic steroids use were recorded. Rhinosinusitis and GER were excluded on the basis of appropriate testing; thus treatment with omalizumab was started when the patient was 9 years old. At age 11, adherence to treatment is satisfactory, with no side effects. More importantly, reduced hospital admissions for asthma exacerbations, no further need for systemic steroids, and improved QoL score (from 6.4 to 6.8) were reported. Finally, progressive step-down of anti-asthma treatment was started, and at present (by 11.5 years) inhaled fluticasone (200 μg/day) plus bimonthly subcutaneous omalizumab provide good control of symptoms. Omalizumab has been continuously administered for 2.6 years and is still ongoing.
A full-term male had severe preschool wheezing and, since age 4, recurrent, severe asthma exacerbations with frequent hospital admissions. At age 8, multiple perennial inhalants and food sensitization (i.e., house dust mites, dog dander, Graminaceae pollen mix, Olea europaea pollen, tomatoes, beans, shrimps, and peas) and high serum IgE levels (1166 KU/l) were found. The patient has been treated over 5 years with inhaled fluticasone (up to 1000 μg/day) in association with salmeterol (100 μg/day) and oral montelukast (5 mg/day). Despite this, monthly hospital admissions with systemic steroids need were recorded. After checking the inhaler technique and adherence to treatment, comorbidities including obesity, rhinosinusitis and GER were excluded. Omalizumab was proposed, but parents refused it. By 13.6 years, despite a treatment including the association of inhaled salmeterol/fluticasone (100 μg/1000 μg∙day − 1 , respectively) plus oral montelukast (10 mg/day), monthly exacerbations requiring systemic steroids are reported.
Discussion and conclusions
Most children and adolescents with asthma respond well to inhaled short-acting beta 2 -agonists (SABA) on demand if symptoms are intermittent, or to low dose controller drugs plus as-needed SABA if the risk of exacerbations increases [ 1 ]. Nevertheless, a proportion of patients is referred to specialists because this strategy is not working and asthma is persistently uncontrolled [ 4 ]. For these children, assessment is primarily aimed at investigating the reasons for poor control. Indeed, when the child is initially referred, before the label of “severe, therapy-resistant asthma” (i.e., not responding to treatment even when factors as exposure to allergens and tobacco smoke have been considered) is assigned, three main categories need to be identified: 1) “not asthma at all”, in which response to treatment is suboptimal because the diagnosis is wrong; 2) “asthma plus ”, when asthma is mild but exacerbated by one or more comorbidities; and 3) “difficult-to-treat asthma”, when asthma is uncontrolled because of potentially reversible factors [ 10 ].
The reported cases highlight some aspects of the disease process that may expand the diagnosis and improve patients’ care. At our institution, the severe asthma program includes a multidisciplinary approach with consultations by gastroenterologists as well as ear, nose and throat experts. Recently, sleep medicine experts joined this multidisciplinary team; thus, unfortunately, sleep-disordered breathing (SDB) could not be excluded at the time of our patients’ assessment. Inhalation technique is periodically evaluated by nurses or doctors in each patient. Unfortunately, in Italy an individual prescription database is not available and thus we cannot assess patients’ use of medication. In two cases, the filtering process eventually identified GER and rhinosinusitis, but poor control of asthma persisted even after comorbidities were treated. In all subjects, inhaler skills, treatment adherence, and environmental exposure to indoor/outdoor allergens as well as to second- and third-hand smoke were excluded as cause of lack of control. Eventually, three out of four patients started anti-IgE treatment; asthma control was obtained and maintenance drugs were progressively reduced. In the case that refused omalizumab therapy, pulmonary function, clinical features and controller treatment including high-dose ICS were unchanged.
Previous studies have highlighted an association between increasing asthma severity in children and reduced QoL [ 11 , 12 , 13 ]. Uncontrolled asthma symptoms not only affect children physically, but can impair them socially, emotionally, and educationally [ 13 ]. In line with previous observations, 3 out 4 of our cases had poor QoL, assessed by a standardized questionnaire [ 14 ]. It is well known that improving QoL in difficult asthma is not an easy task, despite a variety of treatments aimed at achieving control [ 12 ], and much more remains to be done to address the problem. Nevertheless, 2 of our 3 cases showed a remarkable improvement of QoL after one year of treatment with omalizumab.
Reduction in forced expiratory volume in the first second (FEV 1 ) is often used to define childhood asthma severity in treatment guidelines and clinical studies [ 5 , 11 , 15 ]. Nevertheless, children with severe asthma often have a normal FEV 1 that does not improve after bronchodilators, indicating that spirometry may be a poor predictor of asthma severity in childhood [ 6 , 16 , 17 ]. Actually, children with a normal FEV 1 , both before and after β 2 -agonist, may show a bronchodilator response in terms of forced expiratory flow between 25% and 75% (FEF 25–75 ) [ 18 ]. However, the utility of FEF 25–75 in the assessment or treatment of severe asthma is currently unknown. Interestingly, all the reported cases showed normal or slightly reduced values of FEV 1 but severe impairment of FEF 25–75 . Two cases showed a bronchodilator response in terms of FEV 1 (subjects 3 and 4), while 3 patients had a significant increase of FEF 25–75 (cases 1, 3 and 4). Unfortunately, we could not provide the results of bronchodilator response during or after the treatment with omalizumab in any case.
Available literature on the diagnostic approach to difficult asthma in children offers a number of reviews which basically summarize the steps needed to fill the gap between a generic diagnosis of “difficult asthma” and more specific labels (i.e., “severe” asthma, “difficult-to-treat” asthma, or even different diagnoses) [ 3 , 5 , 6 , 8 , 10 , 19 , 20 , 21 ]. So far, few original articles and case reports have been published, probably due to the peculiarity of the issue, which makes retrospective discussion of cases easier than the design of a prospective clinical study [ 4 , 22 , 23 , 24 , 25 , 26 ]. Available knowledge mainly derives from the experience of specialized centers.
The evaluation of a child referred for uncontrolled asthma should start with a careful history focused on typical respiratory symptoms and on the definition of possible triggers. In the “severe asthma” process, it is crucial for clinicians to maintain a high degree of skepticism about the ultimate diagnosis, particularly in the presence of relevant discrepancies between history, physical features and lung function, as many conditions may be misdiagnosed as asthma. In order to simplify this process, herein we propose an algorithm for the diagnosis of difficult-to-treat and severe asthma (Fig. 1 ). Confirmation of the diagnosis through a detailed clinical and laboratory re-evaluation is important because in 12–50% of cases assumed to have severe asthma this might not be the correct diagnosis [ 10 ]. Several documents have indicated the main steps of the process that should be followed in children with uncontrolled asthma [ 3 , 8 , 10 ]. The translation of these procedures into real life practice may deeply change from one subject to another due to the variability of individual patients’ history and clinical features, which will often lead the diagnostic investigations towards the most likely reason for uncontrolled asthma. For children with apparently severe asthma, the first step is to confirm the diagnosis and, before proceeding to broader investigations, to verify that the poor control is not simply determined by poor adherence to treatment, inadequate inhaler skills and/or environmental exposure to triggers. A nurse-led assessment, including a home visit, despite not being applicable in all settings, may be useful for identifying potentially modifiable factors in uncontrolled pediatric asthma [ 27 ].
A practical algorithm for the diagnosis of difficult-to-treat and severe asthma. ICS, inhaled corticosteroids; OCS, oral corticosteroids
A number of comorbidities have been increasingly recognized as factors that may impact asthma clinical expression and control in childhood [ 10 , 28 ]. Children with uncontrolled disease should be investigated for GER, rhinosinusitis, dysfunctional breathing and/or vocal cord dysfunction, obstructive sleep apnea, obesity, psychological factors, smoke exposure, hormonal influences, and ongoing drugs [ 3 , 6 , 8 , 20 ]. Indeed, the exact role played by comorbidities in pediatric asthma control is still debated [ 28 ]. The most impressive example is GER. Several pediatric documents recommend assessing for GER because reflux may be a contributing factor to problematic or difficult asthma [ 7 , 29 ]. Nevertheless, GER treatment might not be effective for severe asthma [ 30 , 31 ], as confirmed by current cases 1 and 2. There is an established evidence that chronic rhinosinusitis is associated with more severe asthma in children [ 32 , 33 , 34 ]. Therefore, examination of upper airways and ad hoc treatment if rhinosinusitis is evident are recommended in children with severe asthma [ 3 , 8 , 35 ]. However, intranasal steroids for rhinitis resulted in a small reduction of asthma risk in school-aged children [ 36 ], and actual placebo-controlled studies on the effect of treatment of rhinosinusitis on asthma control in children are lacking [ 10 , 37 ].
Dysfunctional breathing, including hyperventilation and vocal cord dysfunction, is associated with poorer asthma control in children [ 8 , 10 , 38 , 39 ]. Unfortunately, there is scarce literature on the effect of its treatment on the control of severe asthma in children [ 40 ]. SDB ranging from primary snoring to obstructive sleep apnea syndrome is very common in children [ 41 ], and an increased prevalence of SDB together with increasing asthma severity has been reported [ 42 ]. Interestingly, GER may also be worsened by recurrent episodes of upper airway obstruction associated with SDB, and this may further trigger bronchial obstruction. Asthma guidelines recommend the assessment of SDB through nocturnal polysomnography in poorly controlled asthmatics, particularly if they are also obese [ 5 ]. There are no studies examining whether pediatric asthma improves after SDB has been treated, for example, with nasal steroids, adenotonsillectomy, continuous positive airway pressure or weight reduction if the child is also obese [ 43 ]. The parallel increase in obesity and asthma suggests that the two conditions are linked and that they can aggravate each other [ 44 , 45 ], even though the exact mechanisms that underlie this association remain unclear [ 46 ]. Indeed, other coexisting comorbidities such as SDB or GER may play a confounding role in the development of the interactions between obesity and the airways [ 47 , 48 ]. Obesity is associated with increased markers of inflammation in serum and adipose tissue and yet decreased airway inflammation in obese people with asthma [ 49 ]. Several interventions, including behavioral and weight reduction programs or bariatric surgery, may result in improved asthma control, quality of life and lung function in adult obese asthmatics [ 50 ]. Although reports of adolescent bariatric surgery demonstrate a significant body weight decrease, this approach is not widely available and there are no published reports on its effect on pediatric severe asthma control [ 51 ]. Finally, although it is still unclear whether food allergy is causative or shares a common pathway with difficult asthma, it might explain the loss of asthma control at least in some children and thus be considered as a comorbid condition [ 10 , 16 , 52 ].
In conclusion, establishing the impact of comorbidities on asthma control may be cumbersome, and an ex-juvantibus treatment is sometimes necessary to assess their role. Comorbid conditions can also worsen each other, and symptoms arising from some of them may mimic asthma [ 6 ]. Although the ability to improve pediatric severe asthma by treating comorbidities remains unconfirmed, they should be treated appropriately [ 9 ].
The vast majority of asthmatic children exhibit a mild or at most a moderate disease that can be fully controlled with low-to-medium dose ICS associated or not with other controllers [ 5 , 6 ]. However, a subset of asthmatics remains difficult-to-treat [ 5 , 6 ]. With the advent of biologics, these severe steroid-dependent asthmatics have alternative options for treatment, as steroid-related adverse events are common in severe asthma [ 53 ]. Omalizumab, an anti-IgE monoclonal antibody, is the only biologic therapy recommended in children with moderate-to-severe asthma by the recent guidelines [ 5 , 6 ]. In Italy, this treatment is fully covered by the National Health System. Therefore, there is no influence by any funding on treatment decisions. It was approved by the US (Food and Drug Administration) in 2003 and by the European Union (European Medicines Agency) in 2005 as an add-on treatment for patients aged > 12 years with severe persistent allergic asthma and who have a positive skin test or in-vitro reactivity to a perennial aeroallergen, FEV 1 < 80% predicted, frequent daytime symptoms or nighttime awakenings, and multiple documented severe asthma exacerbations despite daily ICS plus a LABA [ 54 , 55 ]. In 2009, it also received approval in Europe for treating patients aged 6–12 years. Figure 2 illustrates current indications for treatment with omalizumab in children and adolescents with severe asthma.
Indications for omalizumab in children and adolescents with severe asthma
IgE antibodies, Th 2 -derived cytokines and eosinophils play a major role in the development of chronic airway inflammation in asthmatic subjects [ 56 ]. Once released from plasma cells, IgE binds principally to the high-affinity IgE receptor (FcεRI) on mast cells, triggering different effector responses, including the release of mediators leading to allergic inflammatory reactions [ 56 ]. The activation of the allergic cascade by IgE, under constant allergen stimulation, leads to the establishment of chronic allergic inflammation in the airways of asthmatic patients, with IgE being a key element of the vicious circle that maintains it. Cytokines produced during the late phase and subsequent chronic inflammation stage have been directly associated with the induction of airway remodelling, indirectly implicating IgE in the process [ 56 ]. At present, omalizumab is the only commercially available recombinant humanized anti-IgE monoclonal antibody that specifically binds serum free IgE at its CH 3 domain, in the proximity of the binding site for FcεRI, thus preventing IgE from interacting with its receptor on mast cells, basophils, antigen-presenting cells and other inflammatory cells [ 57 ]. The rapid reduction of free IgE levels leads to a downregulation of the FcεRI expression on inflammatory cells and an interruption of the allergic cascade, which results in the reduction of peripheral and bronchial tissue eosinophilia and of levels of granulocyte macrophage colony stimulating factor, interleukin (IL)-2, IL-4, IL-5, and IL-13 [ 58 ]. Moreover, basophils have a relevant role in the initiation and progression of allergic inflammation, suggesting that they may represent a viable therapeutic target. Indeed, in children with severe asthma, it has been reported that omalizumab therapy is associated with a significant reduction in circulating basophil numbers, a finding that is concurrent with improved clinical outcomes [ 59 ]. This finding supports a mechanistic link between IgE levels and circulating basophil populations, and may provide new insights into one mechanism by which omalizumab improves asthma symptoms.
Several clinical controlled and real-life studies of adults with severe, inadequately controlled allergic asthma have demonstrated the efficacy and safety of omalizumab in reducing asthma-related symptoms, corticosteroid use, exacerbation rates, and healthcare resource utilization, and in improving QoL and lung function [ 60 , 61 , 62 , 63 ]. Fewer studies have been published in children. In two double-blind, randomized, placebo-controlled trials (RCTs) of children aged 6 to 12 years with moderate-to-severe allergic asthma, treatment with omalizumab reduced the requirement for ICS and protected against disease exacerbations, but there was little change in asthma symptom scores or spirometry [ 9 , 64 ]. These findings were confirmed and extended in older children [ 65 , 66 , 67 ].
The results of the ICATA study, a multicenter RCT of 419 inner-city children, adolescents and young adults with persistent allergic asthma, showed that, compared to placebo, omalizumab reduces the number of days with asthma symptoms and the proportion of participants with at least one exacerbation by approximately 25% and 19%, respectively ( p < 0.001), thus reducing the need for asthmatic symptom controllers [ 68 ]. Another multicenter RCT of inner-city children and adolescents showed that the addition of omalizumab to ongoing guidelines-based care before patients return to school reduces fall asthma exacerbations (odds ratio, 0.48), particularly in subjects with a recent exacerbation [ 69 ]. Moreover, in a real-life study of 104 children and adolescents with severe allergic refractory asthma followed over 1 year, treatment with omalizumab resulted in good asthma control in 67% of the cases ( p < 0.001), while FEV 1 improved by 4.9% ( p = 0.02) and exacerbation rates and healthcare utilisation decreased approximately by 30% ( p < 0.001) [ 70 ]. The same authors also showed that, after two years of treatment, exacerbation rate and healthcare utilisation were further decreased by 83% and 100%, respectively, while level of asthma control, steroid use and lung function remained unchanged [ 71 ].
A systematic review of pediatric RCTs pooled the data of 1381 children and adolescents with moderate-to-severe allergic asthma in order to establish the efficacy of omalizumab as an add-on therapy [ 72 ]. During the stable-steroid phase, omalizumab decreased the number of patients with at least one exacerbation (risk ratio, 0.69; p < 0.001), the mean number of asthma exacerbations per patient (risk ratio, 0.35; p < 0.001), and the asthma symptom score (mean difference, 0.12; p = 0.005) when compared to placebo. During the steroid reduction phase, omalizumab further reduced the number of patients with at least one exacerbation (risk ratio, 0.48; p < 0.001) and the mean number of asthma exacerbations per patient (mean difference, 0.12; p < 0.05).
Given the cost of omalizumab, many authors have argued for the importance of identifying specific asthma populations who will have significant benefit from it [ 68 , 73 , 74 ]. In the ICATA study, baseline predictors of good response to treatment were sensitization and exposure to cockroach allergen, sensitization to house dust mite allergens, a serum IgE level of more than 100 IU per milliliter, a BMI of 25 or more, and a history of at least one unscheduled medical visit in the previous year [ 68 ].
Several studies have assessed the long-term safety of omalizumab in children and adults. A pooled analysis of 67 RCTs conducted over 2 decades on 4254 children and adults treated with omalizumab showed no association between omalizumab treatment and risk of malignancy [ 75 ]. In an RCT evaluating 225 school-aged children, omalizumab was well tolerated, there were no serious adverse events, and the frequency and types of all adverse events were similar to the placebo group [ 9 ]. These results have been further confirmed by a recent systematic review of RCTs that concluded that treatment with omalizumab does not result in increased risk of malignancy or hypersensitivity reactions [ 72 ].
While the rationale for long-term treatment with omalizumab is supported by pharmacokinetic-pharmacodynamic models [ 76 ], the duration of treatment is still under discussion. Results from published studies suggest that omalizumab should be continued for > 1 year [ 77 , 78 ]. In a retrospective study of adults and children with uncontrolled severe asthma treated with omalizumab, the response to treatment was ‘excellent’ in 52.5% of patients, particularly in the subgroup of children aged 6 to 11 years [ 77 ]. After the discontinuation of treatment, loss of asthma control was documented in 69.2% of the patients who had received omalizumab for < 1 year, 59.1% of the subjects treated for 1–2 years, and 46.1% of the cases treated for > 2 years. Time to loss of control was shorter in younger children and longer in patients with an ‘excellent’ response compared with patients with a ‘good’ response. No early loss of control (within 6 months) was observed among patients with > 3.5 years of continuous treatment with omalizumab. Finally, 20% of patients in whom omalizumab was re-prescribed because of loss of control did not respond to the treatment anymore [ 77 ]. Despite these encouraging findings, the impact of omalizumab on the natural history of severe asthma in children deserves to be further investigated by long-term studies that will also define the criteria and timing for discontinuing the treatment.
It is well known that asthma pharmacotherapy is effective in controlling symptoms and bronchial inflammation, but cannot affect the underlying immune response, thus leading to the possibility of symptom reappearance after its discontinuation [ 79 ]. In this scenario, allergen-specific immunotherapy (AIT) has been proposed as the only therapeutic method that can modulate the underlying immune pathophysiology in allergic asthma [ 80 ].
AIT is currently indicated in children and adults with mild-moderate allergic asthma that is completely or partially controlled by pharmacotherapy and with the evidence of a clear relationship between symptoms and exposure to a specific allergen [ 81 , 82 , 83 , 84 ]. However, according to recent guidelines, the efficacy of AIT in asthmatic subjects is limited, and its potential benefits must be weighed against the risk of side effects and the inconvenience and costs of the prolonged therapy [ 5 ]. Moreover, severe or uncontrolled asthma (regardless of its severity) is a major independent risk factor for non-fatal or even fatal adverse reactions, thus representing a contraindication for AIT [ 85 , 86 , 87 ]. Finally, children with severe asthma are often sensitized to multiple allergens, thus making AIT prescription even more complicated [ 88 ].
In subjects with uncontrolled and/or severe allergic asthma, a combination of omalizumab and AIT has been proposed [ 88 ]. Surprisingly, only a few studies have addressed this issue [ 89 , 90 , 91 , 92 ]. However, pre-treatment with omalizumab seems to improve the efficacy and tolerability of subcutaneous AIT in children and adults with severe allergic asthma both during omalizumab treatment and after its discontinuation [ 89 , 91 , 92 ]. Omalizumab has also been successfully used as a supplementary treatment to AIT in order to improve asthma control in children ≥6 years with severe persistent allergic asthma [ 90 ]. Given the scarcity of studies on AIT plus omalizumab in children with severe allergic asthma, further research is warranted to assess risks and benefits of the combined treatment.
Children with severe asthma require a detailed and individualized approach including re-assessment for differential diagnoses, comorbidities and contributory factors, environmental triggers, lung function and inflammation, adherence and response to therapy, and QoL. Treatment of pediatric severe asthma still relies on the maximal optimal use of corticosteroids, bronchodilators and other controllers recommended for moderate-to-severe disease. However, the management of asthma is becoming much more patient-specific, as more and more is learned about the biology behind the development and progression of asthma.
In the current paper, we described the characteristics of four children with severe asthma in whom omalizumab was prescribed. A review of the relevant literature on the topic was also performed. Finally, we provided an algorithm for the diagnosis of difficult-to-treat and severe asthma in children and adolescents, based on the evidence from the literature review. As all algorithms, it is not meant to replace clinical judgment, but it should drive physicians to adopt a systematic approach towards difficult and severe asthma and provide a useful guide to the clinician.
The addition of omalizumab, the first targeted biological treatment approved for asthma, has led to renewed optimism of outcome improvements in patients with allergic severe asthma. As severe asthma is a heterogeneous condition consisting of different phenotypes, the future of asthma management will likely involve phenotypic and potentially even genotypic characterization in selected cases in order to determine appropriate therapy and thus to provide the highest possible benefit, especially if specific responder phenotypes can be identified and selected for this highly specific treatment.
Abbreviations
Anti-immunoglobulin E
Body mass index
IgE receptor
Forced expiratory flow between 25% and 75%
Forced expiratory volume in the first second
Gastroesophageal reflux
Inhaled corticosteroids
Intensive care unit
Interleukin
Long-acting β 2 -agonist
Oral leukotriene receptor antagonist
Quality of life
Randomized controlled trials
Short-acting β 2 -agonists
Sleep-disordered breathing
O'Byrne PM, Pedersen S, Schatz M, Thoren A, Ekholm E, Carlsson LG, et al. The poorly explored impact of uncontrolled asthma. Chest. 2013;143:511–3.
Article PubMed Google Scholar
National Asthma Education and Prevention Program. Expert panel report 3 (EPR-3): guidelines for the diagnosis and Management of Asthma-Summary Report 2007. J Allergy Clin Immunol. 2007;120:S94–8.
Article Google Scholar
Hedlin G. Management of severe asthma in childhood-state of the art and novel perspectives. Pediatr Allergy Immunol. 2014;25:111–21.
Konradsen JR, Nordlund B, Lidegran M, Pedroletti C, Grönlund H, van Hage M, et al. Problematic severe asthma: a proposed approach to identifying children who are severely resistant to therapy. Pediatr Allergy Immunol. 2011;22:9–18.
Global Initiative for Asthma Report. Global strategy for asthma management and prevention (updated 2016). https://www.ginasthma.org . Accessed 07 June 2017.
Chung KF, Wenzel SE, Brozek JL, Bush A, Castro M, Sterk PJ, et al. International ERS/ATS guidelines on definition, evaluation and treatment of severe asthma. Eur Respir J. 2014;43:343–53.
Article CAS PubMed Google Scholar
Vandenplas Y, Rudolph CD, Di Lorenzo C, Hassall E, Liptak G, Mazur L, et al. Pediatric gastroesophageal reflux clinical practice guidelines: joint recommendations of the north American Society for Pediatric Gastroenterology, Hepatology, and nutrition (NASPGHAN) and the European Society for Pediatric Gastroenterology, Hepatology, and nutrition (ESPGHAN). J Pediatr Gastroenterol Nutr. 2009;49:498–507.
Lødrup Carlsen KC, Hedlin G, Bush A, Wennergren G, de Benedictis FM, De Jongste JC, et al. Assessment of problematic severe asthma in children. Eur Respir J. 2011;37:432–40.
Milgrom H, Berger W, Nayak A, Gupta N, Pollard S, McAlary M, et al. Treatment of childhood asthma with anti-immunoglobulin E antibody (omalizumab). Pediatrics. 2001;108:E36.
Bush A, Saglani S. Management of severe asthma in children. Lancet. 2010;376:814–5.
Article CAS PubMed PubMed Central Google Scholar
Lang A, Mowinckel P, Sachs-Olsen C, Riiser A, Lunde J, Carlsen KH, et al. Asthma severity in childhood, untangling clinical phenotypes. Pediatr Allergy Immunol. 2010;21:945–53.
Nordlund B, Konradsen JR, Pedroletti C, Kull I, Hedlin G. The clinical benefit of evaluating health-related quality-of-life in children with problematic severe asthma. Acta Paediatr. 2011;100:1454–60.
Dean BB, Calimlim BC, Sacco P, Aguilar D, Maykut R, Tinkelman D. Uncontrolled asthma: assessing quality of life and productivity of children and their caregivers using a cross-sectional internet-based survey. Health Qual Life Outcomes. 2010;8:6.
Juniper EF, Guyatt GH, Feeny DH, Ferrie PJ, Griffith LE, Townsend M. Measuring quality of life in children with asthma. Qual Life Res. 1996;5:35–46.
British Thoracic Society. Scottish Intercollegiate Guidelines Network. British guideline on the management of asthma, 2014. https://www.brit-thoracic.org.uk/guidelines-and-quality-standards/asthma-guideline . Accessed 13 Apr 2016.
Montella S, Baraldi E, Cazzato S, Aralla R, Berardi M, Brunetti LM, et al. Severe asthma features in children: a case-control online survey. Ital J Pediatr. 2016;42:9.
Article PubMed PubMed Central Google Scholar
Fitzpatrick AM, Gaston BM, Erzurum SC, Teague WG, National Institutes of Health/National Heart, Lung, and Blood Institute Severe Asthma Research Program. Features of severe asthma in school-age children: Atopy and increased exhaled nitric oxide. J Allergy Clin Immunol. 2006;118:1218–25.
Simon MR, Chinchilli VM, Phillips BR, Sorkness CA, Lemanske RF Jr, Szefler SJ, et al. Forced expiratory flow between 25% and 75% of vital capacity and FEV1/forced vital capacity ratio in relation to clinical and physiological parameters in asthmatic children with normal FEV1 values. J Allergy Clin Immunol. 2010;126:527–34.
Hedlin G, Bush A, Lødrup Carlsen K, Wennergren G, De Benedictis FM, Melén E, et al. Problematic severe asthma in children, not one problem but many: a GA2LEN initiative. Eur Respir J. 2010;36:196–201.
Fitzpatrick AM, Teague WG. Severe asthma in children: insights from the National Heart, Lung, and Blood Institute's severe asthma research program. Pediatr Allergy Immunol Pulmonol. 2010;23:131–8.
Konradsen JR, Caffrey Osvald E, Hedlin G. Update on the current methods for the diagnosis and treatment of severe childhood asthma. Expert Rev Respir Med. 2015;9:769–77.
Lang AM, Konradsen J, Carlsen KH, Sachs-Olsen C, Mowinckel P, Hedlin G, et al. Identifying problematic severe asthma in the individual child—does lung function matter? Acta Paediatr. 2010;99:404–10.
Rao DR, Gaffin JM, Baxi SN, Sheehan WJ, Hoffman EB, Phipatanakul WJ. The utility of forced expiratory flow between 25% and 75% of vital capacity in predicting childhood asthma morbidity and severity. Asthma. 2012;49:586–92.
Eid N, Yandell B, Howell L, Eddy M, Sheikh S. Can peak expiratory flow predict airflow obstruction in children with asthma? Pediatrics. 2000;105:354–8.
Cicutto LC, Chapman KR, Chamberlain D, Downey GP. Difficult asthma: consider all of the possibilities. Can Respir J. 2000;7:415–8.
Wener RR, Bel EH. Severe refractory asthma: an update. Eur Respir Rev. 2013;22:227–35.
Bracken M, Fleming L, Hall P, et al. The importance of nurse-led home visits in the assessment of children with problematic asthma. Arch Dis Child. 2009;94:780–4.
De Groot EP, Kreggemeijer WJ, Brand PL. Getting the basics right resolves most cases of uncontrolled and problematic asthma. Acta Paediatr. 2015;104:916–21.
Grimaldi-Bensouda L, Zureik M, Aubier M, Humbert M, Levy J, Benichou J, et al. Does omalizumab make a difference to the real-life treatment of asthma exacerbations? Results from a large cohort of patients with severe uncontrolled asthma. Chest. 2013;143:398–405.
American Lung Association Asthma Clinical Research Centers, Mastronarde JG, Anthonisen NR, Castro M, Holbrook JT, Leone FT, et al. Efficacy of esomeprazole for treatment of poorly controlled asthma. N Engl J Med. 2009;360:1487–9.
Article PubMed Central Google Scholar
Writing Committee for the American Lung Association Asthma Clinical Research Centers, Holbrook JT, Wise RA, Gold BD, Blake K, Brown ED, et al. Lansoprazole for children with poorly controlled asthma: a randomized controlled trial. JAMA 2012;307:373-381.
Wright AL, Holberg CJ, Martinez FD, Halonen M, Morgan W, Taussig LM. Epidemiology of physician-diagnosed allergic rhinitis in childhood. Pediatrics. 1994;94:895–901.
CAS PubMed Google Scholar
De Groot EP, Nijkamp A, Duiverman EJ, Brand PL. Allergic rhinitis is associated with poor asthma control in children with asthma. Thorax. 2012;67:582–7.
Rotiroti G, Roberts G, Scadding GK. Rhinitis in children: common clinical presentations and differential diagnoses. Pediatr Allergy Immunol. 2015;26:103–10.
Bousquet J, Khaltaev N, Cruz AA, Denburg J, Fokkens WJ, Togias A, et al. Allergic rhinitis and its impact on asthma (ARIA). 2008 update (in collaboration with the World Health Organization, GA(2)LEN and AllerGen). Allergy. 2008;63:S8–160.
Deliu M, Belgrave D, Simpson A, Murray CS, Kerry G, Custovic A. Impact of rhinitis on asthma severity in school-age children. Allergy. 2014;69:1515–21.
Brozek JL, Bousquet J, Baena-Cagnani CE, Bonini S, Canonica GW, Casale TB, et al. Allergic rhinitis and its impact on asthma (ARIA) guidelines: 2010 revision. J Allergy Clin Immunol. 2010;126:466–76.
Weinberger M, Abu-Hasan M. Pseudo-asthma: when cough, wheezing, and dyspnea are not asthma. Pediatrics. 2007;120:855–64.
De Groot EP, Duiverman EJ, Brand PL. Dysfunctional breathing in children with asthma: a rare but relevant comorbidity. Eur Respir J. 2013;41:1068–73.
Barker NJ, Jones M, O'Connell NE, Everard ML. Breathing exercises for dysfunctional breathing/hyperventilation syndrome in children. Cochrane Database Syst Rev. 2013;12:CD010376.
Google Scholar
Section on Pediatric Pulmonology, Subcommittee on Obstructive Sleep Apnea Syndrome, American Academy of Pediatrics. Clinical practice guideline: diagnosis and management of childhood obstructive sleep apnea syndrome. Pediatrics. 2002;109:704–12.
Goldstein NA, Aronin C, Kantrowitz B, Hershcopf R, Fishkin S, Lee H, Weaver DE, et al. The prevalence of sleep-disordered breathing in children with asthma and its behavioral effects. Pediatr Pulmonol. 2015;50:1128–36.
Ross KR, Storfer-Isser A, Hart MA, Kibler AM, Rueschman M, Rosen CL, et al. Sleep-disordered breathing is associated with asthma severity in children. J Pediatr. 2012;160:736–42.
Santamaria F, Montella S, Greco L, Valerio G, Franzese A, Maniscalco M, et al. Obesity duration is associated to pulmonary function impairment in obese subjects. Obesity (Silver Spring). 2011;19:1623–8.
Sivapalan P, Diamant Z, Ulrik CS. Obesity and asthma: current knowledge and future needs. Curr Opin Pulm Med. 2015;21:80–5.
Rasmussen F, Hancox RJ. Mechanisms of obesity in asthma. Curr Opin Allergy Clin Immunol. 2014;14:35–43.
Santamaria F, Montella S, Pietrobelli A. Obesity and pulmonary disease: unanswered questions. Obes Rev. 2012;13:822–33.
Lang JE, Hossain J, Holbrook JT, Teague WG, Gold BD, Wise RA, et al. Gastro-oesophageal reflux and worse asthma control in obese children: a case of symptom misattribution? Thorax. 2016;71:238–46.
Santamaria F, Montella S, De Stefano S, Sperlì F, Barbarano F, Valerio G. Relationship between exhaled nitric oxide and body mass index in children and adolescents. J Allergy Clin Immunol. 2005;116:1163–4.
Van Huisstede A, Rudolphus A, Castro Cabezas M, Biter LU, van de Geijn GJ, Taube C, et al. Effect of bariatric surgery on asthma control, lung function and bronchial and systemic inflammation in morbidly obese subjects with asthma. Thorax. 2015;70:659–67.
Katzmarzyk PT, Bouchard C. Where is the beef? Waist circumference is more highly correlated with BMI and total body fat than with abdominal visceral fat in children. Int J Obes. 2014;38:753–4.
Article CAS Google Scholar
De Groot EP, Duiverman EJ, Brand PL. Comorbidities of asthma during childhood: possibly important, yet poorly studied. Eur Respir J. 2010;36:671–8.
Sweeney J, Patterson CC, Menzies-Gow A, Niven RM, Mansur AH, Bucknall C, et al. Comorbidity in severe asthma requiring systemic corticosteroid therapy: cross-sectional data from the optimum patient care research database and the British thoracic difficult asthma registry. Thorax. 2016; https://doi.org/10.1136/thoraxjnl-2015-207630 .
Federal Drug Administration Advisory for Omalizumab. Available at: https://wayback.archive-it.org/7993/20170111075347/ . http://www.fda.gov/Drugs/DevelopmentApprovalProcess/HowDrugsareDevelopedandApproved/ApprovalApplications/TherapeuticBiologicApplications/default.htm . Accessed 4 Feb 2018.
European Medicines Agency: assessment report for Xolair. Available at: http://www.ema.europa.eu/ema/index.jsp?curl=pages/medicines/human/medicines/000606/human_med_001162.jsp&mid=WC0b01ac058001d124 . Accessed 7 June 2017.
Chung KF. Targeting the interleukin pathway in the treatment of asthma. Lancet. 2015;386:1086–96.
Jensen RK, Plum M, Tjerrild L, Jakob T, Spillner E, Andersen GR. Structure of the omalizumab Fab. Acta Crystallogr F Struct Biol Commun. 2015;71:419–26.
Holgate S, Smith N, Massanari M, Jimenez P. Effects of omalizumab on markers of inflammation in patients with allergic asthma. Allergy. 2009;64:1728–36.
Hill DA, Siracusa MC, Ruymann KR, Tait Wojno ED, Artis D, Spergel JM. Omalizumab therapy is associated with reduced circulating basophil populations in asthmatic children. Allergy. 2014;69:674–7.
Humbert M, Beasley R, Ayres J, Slavin R, Hébert J, Bousquet J, et al. Benefits of omalizumab as add-on therapy in patients with severe persistent asthma who are inadequately controlled despite best available therapy (GINA 2002 step 4 treatment): INNOVATE. Allergy. 2005;60:309–16.
Normansell R, Walker S, Milan SJ, Walters EH, Nair P. Omalizumab for asthma in adults and children. Cochrane Database Syst Rev. 2014;1:CD003559.
Lai T, Wang S, Xu Z, Zhang C, Zhao Y, Hu Y, Cao C, et al. Long-term efficacy and safety of omalizumab in patients with persistent uncontrolled allergic asthma: a systematic review and meta-analysis. Sci Rep. 2015;5:8191.
Abraham I, Alhossan A, Lee CS, Kutbi H, MacDonald K. “real-life” effectiveness studies of omalizumab in adult patients with severe allergic asthma: systematic review. Allergy. 2015; https://doi.org/10.1111/all.12815 .
Lanier B, Bridges T, Kulus M, Taylor AF, Berhane I, Vidaurre CF. Omalizumab for the treatment of exacerbations in children with inadequately controlled allergic (IgE-mediated) asthma. J Allergy Clin Immunol. 2009;124:1210–6.
Solèr M, Matz J, Townley R, Buhl R, O'Brien J, Fox H, et al. The anti-IgE antibody omalizumab reduces exacerbations and steroid requirement in allergic asthmatics. Eur Respir J. 2001;18:254–61.
Holgate ST. Cytokine and anti-cytokine therapy for the treatment of asthma and allergic disease. Cytokine. 2004;28:152–7.
Odajima H, Ebisawa M, Nagakura T, Fujisawa T, Akasawa A, Ito K, et al. Omalizumab in Japanese children with severe allergic asthma uncontrolled with standard therapy. Allergol Int. 2015;64:364–70.
Busse WW, Morgan WJ, Gergen PJ, Mitchell HE, Gern JE, Liu AH, et al. Randomized trial of omalizumab (anti-IgE) for asthma in inner-city children. N Engl J Med. 2011;364:1005–15.
Teach SJ, Gill MA, Togias A, Sorkness CA, Arbes SJ Jr, Calatroni A, et al. Preseasonal treatment with either omalizumab or an inhaled corticosteroid boost to prevent fall asthma exacerbations. J Allergy Clin Immunol. 2015;136:1476–85.
Deschildre A, Marguet C, Salleron J, Pin I, Rittié JL, Derelle J, et al. Add-on omalizumab in children with severe allergic asthma: a 1-year real life survey. Eur Respir J. 2013;42:1224–33.
Deschildre A, Marguet C, Langlois C, Pin I, Rittié JL, Derelle J, et al. Real-life long-term omalizumab therapy in children with severe allergic asthma. Eur Respir J. 2015;46:856–9.
Rodrigo GJ, Neffen H. Systematic review on the use of omalizumab for the treatment of asthmatic children and adolescents. Pediatr Allergy Immunol. 2015;26:551–6.
Oba Y, Salzman GA. Cost-effectiveness analysis of omalizumab in adults and adolescents with moderate-to-severe allergic asthma. J Allergy Clin Immunol. 2004;114:265–9.
Campbell JD, Spackman DE, Sullivan SD. The costs and consequences of omalizumab in uncontrolled asthma from a USA payer perspective. Allergy. 2010;65:1141–8.
Busse W, Buhl R, Fernandez Vidaurre C, Blogg M, Zhu J, Eisner MD, et al. Omalizumab and the risk of malignancy: results from a pooled analysis. J Allergy Clin Immunol. 2012;129:983–9.
Lowe PJ, Renard D. Omalizumab decreases IgE production in patients with allergic (IgE-mediated) asthma; PKPD analysis of a biomarker, total IgE. Br J Clin Pharmacol. 2011;72:306–10.
Molimard M, Mala L, Bourdeix I, Le Gros V. Observational study in severe asthmatic patients after discontinuation of omalizumab for good asthma control. Respir Med. 2014;108:571–6.
Busse WW, Trzaskoma B, Omachi TA, Canvin J, Rosen K, Chipps BE, et al. Evaluating Xolair persistency of response after long-term therapy (XPORT). Am J Respir Crit Care Med. 2014;189:A6576.
Guilbert TW, Morgan WJ, Zeiger RS, Mauger DT, Boehmer SJ, Szefler SJ, et al. Long-term inhaled corticosteroids in preschool children at high risk for asthma. N Engl J Med. 2006;354:1985–97.
Akdis CA. Therapies for allergic inflammation: refining strategies to induce tolerance. Nat Med. 2012;18:736–49.
National Heart, Lung, and Blood Institute. Expert panel report 3: Guidelines for the diagnosis and management of asthma—full report 2007. Available at: https://www.nhlbi.nih.gov/files/docs/guidelines/asthgdln.pdf . Accessed 4 Feb 2018.
Joint Task Force on Practice Parameters, American Academy of Allergy, Asthma and Immunology, American College of Allergy, Asthma and Immunology, Joint Council of Allergy, Asthma and Immunolgy. Allergen immunotherapy: a practice parameter second update. J Allergy Clin Immunol. 2007;120:S25–85.
Zuberbier T, Bachert C, Bousquet PJ, Passalacqua G, Walter Canonica G, Merk H, et al. GA(2) LEN/EAACI pocket guide for allergen-specific immunotherapy for allergic rhinitis and asthma. Allergy. 2010;65:1525–30.
Pajno GB, Bernardini R, Peroni D, Arasi S, Martelli A, Landi M, et al. Clinical practice recommendations for allergen-specific immunotherapy in children: the Italian consensus report. Ital J Pediatr. 2017;43:13.
Pitsios C, Demoly P, Bilo MB, Gerth van Wijk R, Pfaar O, Sturm GJ, et al. Clinical contraindications to allergen immunotherapy: an EAACI position paper. Allergy. 2015;70:897–909.
Tsabouri S, Mavroudi A, Feketea G, Guibas GV. Subcutaneous and sublingual immunotherapy in allergic asthma in children. Front Pediatr. 2017;5:82.
Jutel M, Agache I, Bonini S, Burks AW, Calderon M, Canonica W, et al. International consensus on allergy immunotherapy. J Allergy Clin Immunol. 2015;136:556–68.
Hedlin G, van Hage M. The role of immunotherapy in the management of childhood asthma. Ther Adv Respir Dis. 2012;6:137–46.
Lambert N, Guiddir T, Amat F, Just J. Pre-treatment by omalizumab allows allergen immunotherapy in children and young adults with severe allergic asthma. Pediatr Allergy Immunol. 2014;25:829–32.
Kopp MV, Hamelmann E, Zielen S, Kamin W, Bergmann K-C, Sieder C. Combination of omalizumab and specific immunotherapy is superior to immunotherapy in patients with seasonal allergic rhinoconjunctivitis and co-morbid seasonal allergic asthma. Clin Exp Allergy. 2009;39:271–9.
Massanari M, Nelson H, Casale T, Busse W, Kianifard F, Geba GP. Effect of pretreatment with omalizumab on the tolerability of specific immunotherapy in allergic asthma. J Allergy Clin Immunol. 2010;125:383–9.
Stelmach I, Kaczmarek-Woźniak J, Majak P, Olszowiec-Chlebna M, Jerzynska J. Efficacy and safety of high-doses sublingual immunotherapy in ultra-rush scheme in children allergic to grass pollen. Clin Exp Allergy. 2009;39:401–8.
Download references
Acknowledgements
The authors gratefully thank Dr. Marco Maglione for his contribution in the clinical assessment of the described cases. Medical writing assistance was provided by Stephen Walters on behalf of City Hills Proofreading.
No funding was secured for this study.
Availability of data and materials
All relevant data and materials are published in the manuscript.
Author information
Authors and affiliations.
Department of Translational Medical Sciences, Federico II University, Via Sergio Pansini 5, 80131, Naples, Italy
Virginia Mirra, Silvia Montella & Francesca Santamaria
You can also search for this author in PubMed Google Scholar
Contributions
VM, SM and FS, authors of the current manuscript, declare that they have participated sufficiently in the work to take public responsibility for appropriate portions of the content. VM and SM carried out the initial investigations, drafted the initial manuscript, revised the manuscript, and approved the final manuscript as submitted. FS conceptualized and designed the study, and critically reviewed and approved the final manuscript as submitted. All authors read and approved the final manuscript.
Corresponding author
Correspondence to Francesca Santamaria .
Ethics declarations
Ethics approval and consent to participate.
This study was approved by the ethics committee “Carlo Romano”, Federico II University, Naples, Italy. Children’s parents/legal guardians gave informed written consent to participate. The description of our cases adheres to the CARE standards of reporting checklist.
Consent for publication
Children’s parents/legal guardians provided informed written consent for the case report to be published.
Competing interests
The authors declare that they have no competing interests to disclose. Authors have no financial relationships relevant to this article to disclose.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License ( http://creativecommons.org/licenses/by/4.0/ ), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated.
Reprints and permissions
About this article
Cite this article.
Mirra, V., Montella, S. & Santamaria, F. Pediatric severe asthma: a case series report and perspectives on anti-IgE treatment. BMC Pediatr 18 , 73 (2018). https://doi.org/10.1186/s12887-018-1019-9
Download citation
Received : 24 May 2016
Accepted : 29 January 2018
Published : 21 February 2018
DOI : https://doi.org/10.1186/s12887-018-1019-9
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Severe asthma
- Adolescents
- Asthma exacerbations
BMC Pediatrics
ISSN: 1471-2431
- Submission enquiries: [email protected]
- General enquiries: [email protected]
DBP Community Systems-Based Cases
Introduction.
Following are case studies of children with typical developmental behavioral issues that may require a host of referrals and recommendations.
Case Studies
Case 1: case 2: case 3: sophie mark alejandro.
Neonatal herpes: case series in two obstetric centres over a 10-year period (2013–2023), France
- Published: 27 April 2024
Cite this article

- Elise Bouthry 1 , 2 ,
- Vincent Portet-Sulla 2 , 3 , 4 ,
- Melek Manai Bouokazi 3 ,
- Claire Périllaud-Dubois 2 , 5 ,
- François-Charles Javaugue 3 ,
- Laure Jule 6 ,
- Claire Boithias 6 ,
- Nolwenn Le Saché 6 ,
- Mostafa Mokhtari 6 ,
- Diane Carrière 6 ,
- Louise Sonnier 7 ,
- Rafik Benammar 8 ,
- Alexandra Letourneau 9 ,
- Alexandre J. Vivanti 2 , 9 ,
- Anne-Gaël Cordier 10 , 9 nAff2 ,
- Emmanuelle Letamendia-Richard 8 &
- Christelle Vauloup-Fellous 2 , 3 , 4
32 Accesses
1 Altmetric
Explore all metrics
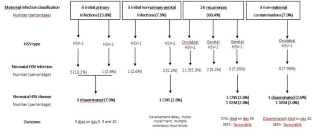
Neonatal herpes simplex virus (HSV) infection (HSV infection in infants less than 6 weeks of age) is rare but mortality and morbidity rates are high after disseminated disease and encephalitis. In France, the epidemiology is poorly described, and two decades ago, incidence was estimated to be 3 per 100,000 live births a year. We describe determinants, epidemiologic and clinical characteristics of neonatal HSV infection in a managed-care population attending in two major obstetric and paediatric centres, Paris, France, over a 10-year period. This retrospective case series study was conducted from 2013 to 2023, in infants less than 42 days of age who had virologically confirmed HSV infection. We report an overall rate of neonatal herpes of 5.5 per 100,000 live births a year and an incidence of symptomatic cases of 1.2 per 100,000 live births a year. HSV-1 was the major serotype involved (84.2%) and post-natal acquisition through the orolabial route reached 63.2%. All neonates who had neonatal HSV PCR screening (owing to clinical signs in parents) and who received prompt acyclovir treatment remained asymptomatic. Symptomatic forms accounted for 21.1% cases of the total and mortality was high (62.5% of symptomatic forms).
Conclusion : This case series confirms that neonates at risk for HSV disease and poor outcome are those born to HSV-seronegative mothers, preterm infants, and those who received acyclovir after onset of symptoms (mainly because mothers did not present evidence of acute HSV infection). Our study confirms the major role of HSV-1 and the frequency of its early post-natal acquisition.
This is a preview of subscription content, log in via an institution to check access.
Access this article
Price includes VAT (Russian Federation)
Instant access to the full article PDF.
Rent this article via DeepDyve
Institutional subscriptions
Similar content being viewed by others
Childhood community-acquired pneumonia
Viral meningitis: an overview
Neurological Complications of Dengue Fever
Data availability.
All data supporting the findings of this study are available within the paper and its supplementary information.
Abbreviations
Central nervous disease
Cycle threshold
- Herpes simplex virus
Initial non-primary infection
Initial primary infection
Polymerase chain reaction
Skin, eyes, mouth
Week of gestation
Looker KJ, Magaret AS, May MT, Turner KME, Vickerman P, Newman L, Gottlieb S (2017) First estimates of the global and regional incidence of neonatal herpes infection. Lancet Glob Health 5:e300–e309. https://doi.org/10.1016/S2214-109X(16)30362-X
Article PubMed PubMed Central Google Scholar
Bernstein DI, Bellamy AR, Hook EW, Levin MJ, Wald A, Ewell MG, Wolff PA, Deal CD, Heineman TC, Dubin G, Belshe RB (2013) Epidemiology, clinical presentation, and antibody response to primary infection with herpes simplex virus type 1 and type 2 in young women. Clin Infect Dis 56:344–351. https://doi.org/10.1093/cid/cis891
Article CAS PubMed Google Scholar
Looker KJ, Magaret AS, Turner KME, Vickerman P, Gottlieb S, Newman L (2015) Global estimates of prevalent and incident herpes simplex virus type 2 infections in 2012. PLoS ONE 10. https://doi.org/10.1371/journal.pone.0114989
Article CAS PubMed PubMed Central Google Scholar
Delaney S, Gardella C, Saracino M, Margaret A, Wald A (2014) Seroprevalence of herpes simplex virus type 1 and 2 among pregnant women, 1989–2010. Obstet Gynecol Surv 312:746–748. https://doi.org/10.1001/jama.2014.4359
Article CAS Google Scholar
Looker KJ, Magaret AS, May MT, Turner KME, Vickerman P, Gottlieb S, Newman L (2015) Global and regional estimates of prevalent and incident herpes simplex virus type 1 infections in 2012. PLoS ONE 10. https://doi.org/10.1371/journal.pone.0140765
Picone O (2017) Genital herpes and pregnancy: epidemiology, clinical manifestations, prevention and screening. Guidelines for clinical practice from the French College of Gynecologists and Obstetrician (CNGOF). Gynecol Obstet Fertil Senol 45:642–654
CAS PubMed Google Scholar
Brown ZA, Selke S, Zeh J, Kopelman J, Maslow A, Ashley RL, Watts DH, Berry S, Herd M, Corey L (1997) The acquisition of herpes simplex virus during pregnancy. N Engl J Med 337:509–515. https://doi.org/10.1056/NEJM199708213370801
Gupta R, Warren T, Wald A (2007) Genital herpes Lancet Lond Engl 370:2127–2137. https://doi.org/10.1016/S0140-6736(07)61908-4
Article Google Scholar
Aujard Y (2002) Modalities of treatment local and general, medicamentous or not, controlling neonate suspected to be infected/contaminated by HSV1 or HSV2. Ann Dermatol Venereol 129:655–661
Garland SM, Steben M (2014) Genital herpes. Best Pract Res Clin Obstet Gynaecol 28:1098–1110. https://doi.org/10.1016/j.bpobgyn.2014.07.015
Article PubMed Google Scholar
Brown ZA, Benedetti J, Ashley R, Burchett S, Selke S, Berry S, Vontver LA, Corey L (1991) Neonatal herpes simplex virus infection in relation to asymptomatic maternal infection at the time of labor. N Engl J Med 324:1247–1252. https://doi.org/10.1056/NEJM199105023241804
Whitley R, Arvin A, Prober C, Corey L, Burchett S, Plotkin S, Starr S, Jacobs R, Powell D, Nahmias A et al (1991) Predictors of morbidity and mortality in neonates with herpes simplex virus infections. The National Institute of Allergy and Infectious Diseases Collaborative Antiviral Study Group. N Engl J Med 14(324):450–454. https://doi.org/10.1056/NEJM199102143240704
Kimberlin DW, Lin CY, Jacobs RF, Powell DA, Frenkel LM, Gruber WC, Rathore M, Bradley JS, Diaz PS, Kumar M, Arvin AM et al (2001) Natural history of neonatal herpes simplex virus infections in the acyclovir era. Pediatrics 108:223–229. https://doi.org/10.1542/peds.108.2.223
Caviness AC, Demmler GJ, Selwyn BJ (2008) Clinical and laboratory features of neonatal herpes simplex virus infection: a case-control study. Pediatr Infect Dis J 27:425–430. https://doi.org/10.1097/INF.0b013e3181646d95
Morris SR, Bauer HM, Samuel MC, Gallagher D, Bolan G (2008) Neonatal herpes morbidity and mortality in California, 1995–2003. Sex Transm Dis 35:14–18
Kropp RY, Wong T, Cormier L, Ringrose A, Burton S, Embree JE, Steben M (2006) Neonatal herpes simplex virus infections in Canada: results of a 3-year national prospective study. Pediatrics 117:1955–1962. https://doi.org/10.1542/peds.2005-1778
Jones CA, Raynes-Greenow C, Isaacs D (2014) Population-based surveillance of neonatal herpes simplex virus infection in Australia, 1997–2011. Clin Infect Dis 59:525–531. https://doi.org/10.1093/cid/ciu381
Koren A, Tasher D, Stein M, Yossepowitch O, Somekh E (2013) Neonatal herpes simplex virus infections in Israel. Pediatr Infect Dis J 32:120–123. https://doi.org/10.1097/INF.0b013e3182717f0b
Hemelaar SJAL, Poeran J, Steegers EAP, van der Meijden WI (2015) Neonatal herpes infections in the Netherlands in the period 2006–2011. J Matern Fetal Neonatal Med 28:905–909. https://doi.org/10.3109/14767058.2014.937691
Curfman AL, Glissmeyer EW, Ahmad FA, Korgenski EK, Blaschke AJ, Byington CL, Miller AS (2016) Initial presentation of neonatal herpes simplex virus infection. J Pediatr 172:121-126.e1. https://doi.org/10.1016/j.jpeds.2016.02.015
Lopez-Medina E, Cantey JB, Sánchez PJ (2015) The mortality of neonatal herpes simplex virus infection. J Pediatr 166:1529-1532.e1. https://doi.org/10.1016/j.jpeds.2015.03.004
Kimberlin DW, Whitley RJ, Wan W, Powell DA, Storch G, Ahmed A, Palmer A, Sánchez PJ, Jacobs RF, Bradley JS et al (2011) Oral acyclovir suppression and neurodevelopment after neonatal herpes. N Engl J Med 365:1284–1292. https://doi.org/10.1056/NEJMoa1003509
Shane AL, Sánchez PJ, Stoll BJ (2017) Neonatal sepsis. Lancet 390:1770–1780. https://doi.org/10.1016/S0140-6736(17)31002-4
Gantt S, Muller WJ (2013) The immunologic basis for severe neonatal herpes disease and potential strategies for therapeutic intervention. Clin Dev Immunol 2013. https://doi.org/10.1155/2013/369172
Renesme L (2017) Neonatal herpes: epidemiology, clinical manifestations and management. Guidelines for clinical practice from the French College of Gynecologists and Obstetricians (CNGOF). Gynecol Obstet Fertil Senol 45:691–704. https://doi.org/10.1016/j.gofs.2017.10.005
Kimberlin DW, Baley J (2013) Guidance on management of asymptomatic neonates born to women with active genital herpes lesions. Pediatrics 131:e635-646. https://doi.org/10.1542/peds.2012-3216
Corey L, Wald A (2009) Maternal and neonatal herpes simplex virus infections. N Engl J Med 361:1376–1378. https://doi.org/10.1056/NEJMra0807633
Brown ZA, Wald A, Morrow RA, Selke S, Zeh J, Corey L (2003) Effect of serologic status and cesarean delivery on transmission tates of herpes simplex virus from mother to infant. JAMA 289:203–209. https://doi.org/10.1001/jama.289.2.203
Whitley R, Davis EA, Suppapanya N (2007) Incidence of neonatal herpes simplex virus infections in a managed-care population. Sex Transm Dis 34:704–708. https://doi.org/10.1097/01.olq.0000258432.33412.e2
Batra D, Davies P, Manktelow BN, Smith C (2014) The incidence and presentation of neonatal herpes in a single UK tertiary centre, 2006–2013. Arch Dis Child Educ Pract Ed 99:916–921. https://doi.org/10.1136/archdischild-2013-305335
Pascual A, Moessinger A, Gerber S, Meylan P, Swiss Paediatric Surveillance Unit (SPSU) (2011) l. Neonatal herpes simplex virus infections in Switzerland: results of a 6-year national prospective surveillance study. Clin Microbiol Infect 17:1907–1910. https://doi.org/10.1111/j.1469-0691.2011.03641.x
Flagg EW, Weinstock H (2011) Incidence of neonatal herpes simplex virus infections in the United States, 2006. Pediatrics 127:e1-8. https://doi.org/10.1542/peds.2010-0134
Aymard M, Braig S, Judlin P, F et al (2021) Herpès néonatal : enquête rétrospective française (1998–1999). Arch Pédiatr 8:549
Google Scholar
Cantey JB, Klein AM, Sánchez PJ (2015) Herpes simplex virus DNAemia preceding neonatal disease. J Pediatr 166:1308–1309. https://doi.org/10.1016/j.jpeds.2015.01.042
Kimberlin DW, Lakeman FD, Arvin AM, Prober CG, Corey L, Powell DA, Burchett SK, Jacobs RF, Starr SE, Whitley RJ (1996) Application of the polymerase chain reaction to the diagnosis and management of neonatal herpes simplex virus disease. J Infect Dis 174:1162–1167. https://doi.org/10.1093/infdis/174.6.1162
Cantey JB, Mejías A, Wallihan R, Doern C, Brock E, Salamon D, Marcon M, Sánchez PJ (2012) Use of blood polymerase chain reaction testing for diagnosis of herpes simplex virus infection. J Pediatr 161:357–361. https://doi.org/10.1016/j.jpeds.2012.04.009
Chua C, Arnolds M, Niklas V (2015) Molecular diagnostics and newborns at risk for genital herpes simplex virus. Pediatr Ann 44:e97-102. https://doi.org/10.3928/00904481-20150512-08
Melvin AJ, Mohan KM, Schiffer JT, Drolette LM, Magaret A, Corey L, Wald A (2015) Plasma and cerebrospinal fluid herpes simplex virus levels at diagnosis and outcome of neonatal infection. J Pediatr 166:827–833. https://doi.org/10.1016/j.jpeds.2014.11.011
Brower LH, Wilson PM, Kurowski EM, Haslam D, Courter J, Goyal N, Durling M, Shah SS, Schondelmeyer A (2019) Using quality improvement to implement a standardized approach to neonatal herpes simplex virus. Pediatrics. https://doi.org/10.1542/PEDS.2018-0262
O’Riordan DP, Golden WC, Aucott SW (2006) Herpes simplex virus infections in preterm infants. Pediatrics 118:e1612–e1622. https://doi.org/10.1542/peds.2005-1228
van der Wiel H, Weiland HT, van Doornum GJ, van der Straaten PJ, Berger HM (1985) Disseminated neonatal herpes simplex virus infection acquired from the father. Eur J Pediatr 144:56–57. https://doi.org/10.1007/BF00491927
Yeager AS, Ashley RL, Corey L (1983) Transmission of herpes simplex virus from father to neonate. J Pediatr 103:905–907. https://doi.org/10.1016/s0022-3476(83)80710-0
Bal A, Zandotti C, Nougairede A, Ninove L, Roquelaure B, Charrel RN (2013) Fulminant hepatitis due to father-to-newborn transmission of herpes simplex virus type 1. Open Virol J 7:96–97. https://doi.org/10.2174/1874357901307010096
Ramchandani M, Kong M, Tronstein E, Selke S, Mikhaylova A, Magaret A, Huang ML, Johnston C, Corey L, Wald A (2016) Herpes simplex virus type 1 shedding in tears and nasal and oral mucosa of healthy adults. Sex Transm Dis 43:756–760. https://doi.org/10.1097/OLQ.0000000000000522
Hammerberg O, Watts J, Chernesky M, Luchsinger I, Rawls W (1983) An outbreak of herpes simplex virus type 1 in an intensive care nursery. Pediatr Infect Dis 2:290–294. https://doi.org/10.1097/00006454-198307000-00007
Linnemann CC, Buchman TG, Light IJ, Ballard JL (1978) Transmission of herpes-simplex virus type 1 in a nursery for the newborn. Identification of viral isolates by D.N.A. “fingerprinting.” Lancet 1:964–966. https://doi.org/10.1016/s0140-6736(78)90251-9
Pinninti SG, Kimberlin DW (2013) Maternal and neonatal herpes simplex virus infections. Am J Perinatol 30:113–119. https://doi.org/10.1055/s-0032-1332802
De Guzman MC, Chawla R, Duttchen K (2014) Possible neonatal herpes simplex virus (HSV) acquired postpartum from maternal oral HSV reactivation after neuraxial morphine. A A Case Rep 2:103–105. https://doi.org/10.1213/XAA.0000000000000013
Van Der Wiel H, Weiland HT, Van Doornum GJJ, van der Straaten PJ, H M Berger HM (1985) Disseminated neonatal herpes simplex virus infection acquired from the father. Eur J Pediatr 144:56–57. https://doi.org/10.1007/BF00491927
Bradley H, Markowitz LE, Gibson T, McQuillan GM (2014) Seroprevalence of herpes simplex virus types 1 and 2-United States, 1999–2010. J Infect Dis 209:325–333. https://doi.org/10.1093/infdis/jit458
Mercer CH, Tanton C, Prah P, Erens B, Sonnenberg P, Clifton S, Macdowall W, Lewis R, Field N, Datta J et al (2013) Changes in sexual attitudes and lifestyles in Britain through the life course and over time: findings from the National Surveys of Sexual Attitudes and Lifestyles (Natsal). Lancet 382:1781–1794. https://doi.org/10.1016/S0140-6736(13)62035-8
Gnann JW, Whitley RJ (2016) Clinical practice. Genital Herpes N Engl J Med 375:666–674. https://doi.org/10.1056/NEJMcp1603178
Gupta R, Warren T, Wald A (2007) Genital herpes. Lancet 370:2127–2133. https://doi.org/10.1016/S0140-6736(07)61908-4
Langenberg AG, Corey L, Ashley RL, Leong WP, Straus SE (1999) A prospective study of new infections with herpes simplex virus type 1 and type 2. Chiron HSV Vaccine Study Group. N Engl J Med 341:1432–1438. https://doi.org/10.1056/NEJM199911043411904
Kidszun A, Bruns A, Schreiner D, TippmannS WJ, Pokora RM, Urschitz MS, Mildenberger E (2022) Characteristics of neonatal herpes simplex virus infections in Germany: results of a 2-year prospective nationwide surveillance study. Arch Dis Child Fetal Neonatal Ed 107:F188–F192. https://doi.org/10.1136/ARCHDISCHILD-2021-321940
Dungu KHS, Lund S, Malchau Carlsen EL, Hartling UL, Matthesen AT, Franck KT, MK, Ulrik Justesen S, Nielsen HL, Nielsen ALY, et al (2023) Herpes simplex virus infection among neonates suspected of invasive bacterial infection: a population-based cohort study. Arch Dis Child Fetal Neonatal. https://doi.org/10.1136/ARCHDISCHILD-2023-325583
Teutsch SM, Nunez CA, Morris A, Eslick GD, Berkhout A, Novakovic D, Brotherton JML, McGregor S, Khawar L, Khandaker G et al (2022) Australian Paediatric Surveillance Unit (APSU) Annual Surveillance Report 2021. Commun Dis Intell. https://doi.org/10.33321/CDI.2022.46.66
Matthias J, Du Bernard S, Schillinger JA, Hong J, Pearson V, Peterman TA (2021) Estimating neonatal herpes simplex virus incidence and mortality using capture-recapture, Florida. Clin Infect Dis 73:506–512. https://doi.org/10.1093/CID/CIAA727
Kimberlin DW (2014) The scarlet H. J Infect Dis 209:315–317. https://doi.org/10.1093/infdis/jit459
Download references
Author information
Anne-Gaël Cordier
Present address: Groupe de Recherche sur les Infections pendant la grossesse (GRIG), Paris, France
Authors and Affiliations
Department of Virology, Angers University Hospital, 4 rue Larrey, 49933, Angers, France
Elise Bouthry
Groupe de Recherche sur les Infections pendant la grossesse (GRIG), Paris, France
Elise Bouthry, Vincent Portet-Sulla, Claire Périllaud-Dubois, Alexandre J. Vivanti & Christelle Vauloup-Fellous
Division of Virology, WHO Rubella National Reference Laboratory, Dept of Biology and Genetics, Paris Saclay University Hospital, APHP, Paris, France
Vincent Portet-Sulla, Melek Manai Bouokazi, François-Charles Javaugue & Christelle Vauloup-Fellous
Center for Immunology of Viral, Hematological and Bacterial Diseases (IMVA-HB/IDMIT), Université Paris-Saclay, INSERM U1184, CEA, Auto-Immune, Fontenay-Aux-Roses, France
Vincent Portet-Sulla & Christelle Vauloup-Fellous
Virology Department, Sorbonne University, Saint-Antoine Hospital, AP-HP, Pierre Louis Epidemiology and Public Health Institute (iPLESP), INSERM 1136, Paris, France
Claire Périllaud-Dubois
Division of Paediatric Critical Care and Neonatal Medicine, FAME Department, Paris Saclay University Hospital, “Kremlin-Bicetre” Medical Center-APHP, Paris, France
Laure Jule, Claire Boithias, Nolwenn Le Saché, Mostafa Mokhtari & Diane Carrière
Division of Obstetrics and Gynecology, Centre Hospitalier Simone Veil, 41000, Blois, France
Louise Sonnier
Department of Neonatal Medicine, DMU2 Santé des Femmes et des Nouveau-nés, Paris Saclay University Hospital, “Antoine Béclère” Medical Center-APHP, Paris, France
Rafik Benammar & Emmanuelle Letamendia-Richard
Division of Obstetrics and Gynecology, DMU Santé des Femmes et des Nouveau-Nés, Paris Saclay University Hospital, “Antoine Béclère” Medical Center-APHP, Paris, France
Alexandra Letourneau, Alexandre J. Vivanti & Anne-Gaël Cordier
Department of Obstetrics and Gynecology, AP-HP.Sorbonne Université, “Tenon” Medical Center-APHP, Paris, France
You can also search for this author in PubMed Google Scholar
Contributions
FC.J and C.VF designed and implemented the study. E.B, V.PS, C.PD, FC.J, M.MB, and C.VF performed and interpreted routine virological analysis and, if necessary, complementary analysis. L.J, C.B, N.LS, M.M, D.C, L.S, R.B, A.L, AJ.V, AG.C, and E.L managed the patients and their mothers. E.B, FC.J, M.MB, and C.VF collected the data. FC.J and C.VF analyzed and interpreted the data, and drafted the manuscript. All authors critically reviewed the manuscript for important intellectual content and approved the final version. All authors agree to be accountable for all aspects of this work.
Corresponding author
Correspondence to Elise Bouthry .
Ethics declarations
Conflict of interest.
The authors declare no competing interests.
Additional information
Communicated by Tobias Tenenbaum
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 (DOC 114 KB)
Rights and permissions.
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Reprints and permissions
About this article
Bouthry, E., Portet-Sulla, V., Bouokazi, M.M. et al. Neonatal herpes: case series in two obstetric centres over a 10-year period (2013–2023), France. Eur J Pediatr (2024). https://doi.org/10.1007/s00431-024-05581-9
Download citation
Received : 20 February 2024
Revised : 15 April 2024
Accepted : 19 April 2024
Published : 27 April 2024
DOI : https://doi.org/10.1007/s00431-024-05581-9
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Neonatal herpes
- Maternal infection
- Find a journal
- Publish with us
- Track your research
- Advanced search

Advanced Search
A Master Protocol Template for Pediatric ARDS Studies
- Find this author on Google Scholar
- Find this author on PubMed
- Search for this author on this site
- For correspondence: [email protected]
- Info & Metrics
BACKGROUND: Pediatric ARDS is associated with significant morbidity and mortality. High-quality data from clinical trials in children are limited due to numerous barriers to their design and execution. Here we describe the collaborative development of a master protocol as a tool to address some of these barriers and support the conduct of pediatric ARDS studies.
METHODS: Using PubMed, we performed a literature search of randomized controlled trials (RCTs) in pediatric ARDS to characterize the current state and evaluate potential benefit of harmonized master protocols. We used a multi-stakeholder, collaborative, and team science–oriented process to develop a master protocol template with links to common data elements (CDEs) for pediatric ARDS trials.
RESULTS: We identified 11 RCTs that enrolled between 14–200 total subjects per trial. Interventions included mechanical ventilation, prone positioning, corticosteroids, and surfactant. Studies displayed significant heterogeneity in ARDS definition, design, inclusion and exclusion criteria, and reported outcomes. Mortality was reported in 91% of trials and ventilator-free days in 73%. The trial heterogeneity made pooled analysis unfeasible. These findings underscore the need for a method to facilitate combined analysis of future trials through standardization of trial elements. As a potential solution, we developed a master protocol, iteratively revised with input from a multidisciplinary panel of experts and organized into 3 categories: instructions and general information, templated language, and a series of text options of common pediatric ARDS trial scenarios. Finally, we linked master protocol sections to relevant CDEs previously defined for pediatric ARDS and captured in a series of electronic case report forms.
CONCLUSIONS: The majority of pediatric ARDS trials identified were small and heterogeneous in study design and outcome reporting. Using a master protocol template for pediatric ARDS trials with CDEs would support combining and comparing pediatric ARDS trial findings and increase the knowledge base.
- mechanical ventilation
- clinical trial
- Correspondence: Andrew G Miller MSc RRT RRT-NPS RRT-ACCS FAARC, 2301 Erwin Road, Durham, NC, 27710 . E-mail: Andrew.g.miller{at}duke.edu
- Copyright © 2024 by Daedalus Enterprises
In this issue

- Table of Contents
- Table of Contents (PDF)
- Cover (PDF)
- Index by author
Thank you for your interest in spreading the word on American Association for Respiratory Care.
NOTE: We only request your email address so that the person you are recommending the page to knows that you wanted them to see it, and that it is not junk mail. We do not capture any email address.
Citation Manager Formats
- EndNote (tagged)
- EndNote 8 (xml)
- RefWorks Tagged
- Ref Manager

- Tweet Widget
- Facebook Like
- Google Plus One
Jump to section
Related articles, cited by....
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 01 May 2024
A critical assessment of using ChatGPT for extracting structured data from clinical notes
- Jingwei Huang ORCID: orcid.org/0000-0003-2155-6107 1 ,
- Donghan M. Yang 1 ,
- Ruichen Rong 1 ,
- Kuroush Nezafati ORCID: orcid.org/0000-0002-6785-7362 1 ,
- Colin Treager 1 ,
- Zhikai Chi ORCID: orcid.org/0000-0002-3601-3351 2 ,
- Shidan Wang ORCID: orcid.org/0000-0002-0001-3261 1 ,
- Xian Cheng 1 ,
- Yujia Guo 1 ,
- Laura J. Klesse 3 ,
- Guanghua Xiao 1 ,
- Eric D. Peterson 4 ,
- Xiaowei Zhan 1 &
- Yang Xie ORCID: orcid.org/0000-0001-9456-1762 1
npj Digital Medicine volume 7 , Article number: 106 ( 2024 ) Cite this article
38 Altmetric
Metrics details
- Non-small-cell lung cancer
Existing natural language processing (NLP) methods to convert free-text clinical notes into structured data often require problem-specific annotations and model training. This study aims to evaluate ChatGPT’s capacity to extract information from free-text medical notes efficiently and comprehensively. We developed a large language model (LLM)-based workflow, utilizing systems engineering methodology and spiral “prompt engineering” process, leveraging OpenAI’s API for batch querying ChatGPT. We evaluated the effectiveness of this method using a dataset of more than 1000 lung cancer pathology reports and a dataset of 191 pediatric osteosarcoma pathology reports, comparing the ChatGPT-3.5 (gpt-3.5-turbo-16k) outputs with expert-curated structured data. ChatGPT-3.5 demonstrated the ability to extract pathological classifications with an overall accuracy of 89%, in lung cancer dataset, outperforming the performance of two traditional NLP methods. The performance is influenced by the design of the instructive prompt. Our case analysis shows that most misclassifications were due to the lack of highly specialized pathology terminology, and erroneous interpretation of TNM staging rules. Reproducibility shows the relatively stable performance of ChatGPT-3.5 over time. In pediatric osteosarcoma dataset, ChatGPT-3.5 accurately classified both grades and margin status with accuracy of 98.6% and 100% respectively. Our study shows the feasibility of using ChatGPT to process large volumes of clinical notes for structured information extraction without requiring extensive task-specific human annotation and model training. The results underscore the potential role of LLMs in transforming unstructured healthcare data into structured formats, thereby supporting research and aiding clinical decision-making.
Similar content being viewed by others

Systematic analysis of ChatGPT, Google search and Llama 2 for clinical decision support tasks
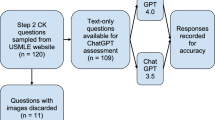
Assessing ChatGPT 4.0’s test performance and clinical diagnostic accuracy on USMLE STEP 2 CK and clinical case reports
Large language models streamline automated machine learning for clinical studies
Introduction.
Large Language Models (LLMs) 1 , 2 , 3 , 4 , 5 , 6 , such as Generative Pre-trained Transformer (GPT) models represented by ChatGPT, are being utilized for diverse applications across various sectors. In the healthcare industry, early applications of LLMs are being used to facilitate patient-clinician communication 7 , 8 . To date, few studies have examined the potential of LLMs in reading and interpreting clinical notes, turning unstructured texts into structured, analyzable data.
Traditionally, the automated extraction of structured data elements from medical notes has relied on medical natural language processing (NLP) using rule-based or machine-learning approaches or a combination of both 9 , 10 . Machine learning methods 11 , 12 , 13 , 14 , particularly deep learning, typically employ neural networks and the first generation of transformer-based large language models (e.g., BERT). Medical domain knowledge needs to be integrated into model designs to enhance performance. However, a significant obstacle to developing these traditional medical NLP algorithms is the limited existence of human-annotated datasets and the costs associated with new human annotation 15 . Despite meticulous ground-truth labeling, the relatively small corpus sizes often result in models with poor generalizability or make evaluations of generalizability impossible. For decades, conventional artificial intelligence (AI) systems (symbolic and neural networks) have suffered from a lack of general knowledge and commonsense reasoning. LLMs, like GPT, offer a promising alternative, potentially using commonsense reasoning and broad general knowledge to facilitate language processing.
ChatGPT is the application interface of the GPT model family. This study explores an approach to using ChatGPT to extract structured data elements from unstructured clinical notes. In this study, we selected lung cancer pathology reports as the corpus for extracting detailed diagnosis information for lung cancer. To accomplish this, we developed and improved a prompt engineering process. We then evaluated the effectiveness of this method by comparing the ChatGPT output with expert-curated structured data and used case studies to provide insights into how ChatGPT read and interpreted notes and why it made mistakes in some cases.
Data and endpoints
The primary objective of this study was to develop an algorithm and assess the capabilities of ChatGPT in processing and interpreting a large volume of free-text clinical notes. To evaluate this, we utilized unstructured lung cancer pathology notes, which provide diagnostic information essential for developing treatment plans and play vital roles in clinical and translational research. We accessed a total of 1026 lung cancer pathology reports from two web portals: the Cancer Digital Slide Archive (CDSA data) ( https://cancer.digitalslidearchive.org/ ) and The Cancer Genome Atlas (TCGA data) ( https://cBioPortal.org ). These platforms serve as public data repositories for de-identified patient information, facilitating cancer research. The CDSA dataset was utilized as the “training” data for prompt development, while the TCGA dataset, after removing the overlapping cases with CDSA, served as the test data for evaluating the ChatGPT model performance.
From all the downloaded 99 pathology reports from CDSA for the training data, we excluded 21 invalid reports due to near-empty content, poor scanning quality, or missing report forms. Seventy-eight valid pathology reports were included as the training data to optimize the prompt. To evaluate the model performance, 1024 pathology reports were downloaded from cBioPortal. Among them, 97 overlapped with the training data and were excluded from the evaluation. We further excluded 153 invalid reports due to near-empty content, poor scanning quality, or missing report forms. The invalid reports were preserved to evaluate ChatGPT’s handling of irregular inputs separately, and were not included in the testing data for accuracy performance assessment. As a result, 774 valid pathology reports were included as the testing data for performance evaluation. These valid reports still contain typos, missing words, random characters, incomplete contents, and other quality issues challenging human reading. The corresponding numbers of reports used at each step of the process are detailed in Fig. 1 .
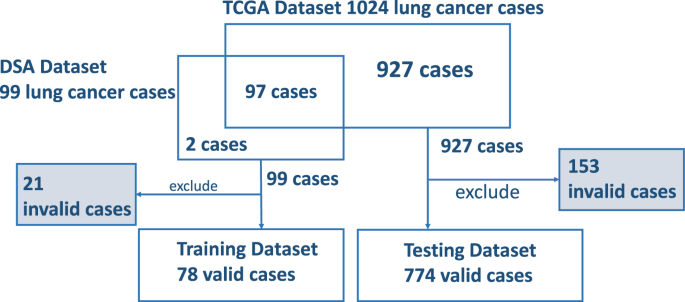
Exclusions are accounted for due to reasons such as empty reports, poor scanning quality, and other factors, including reports of stage IV or unknown conditions.
The specific task of this study was to identify tumor staging and histology types which are important for clinical care and research from pathology reports. The TNM staging system 16 , outlining the primary tumor features (T), regional lymph node involvement (N), and distant metastases (M), is commonly used to define the disease extent, assign prognosis, and guide lung cancer treatment. The American Joint Committee on Cancer (AJCC) has periodically released various editions 16 of TNM classification/staging for lung cancers based on recommendations from extensive database analyses. Following the AJCC guideline, individual pathologic T, N, and M stage components can be summarized into an overall pathologic staging score of Stage I, II, III, or IV. For this project, we instructed ChatGPT to use the AJCC 7 th edition Cancer Staging Manual 17 as the reference for staging lung cancer cases. As the lung cancer cases in our dataset are predominantly non-metastatic, the pathologic metastasis (pM) stage was not extracted. The data elements we chose to extract and evaluate for this study are pathologic primary tumor (pT) and pathologic lymph node (pN) stage components, overall pathologic tumor stage, and histology type.
Overall Performance
Using the training data in the CDSA dataset ( n = 78), we experimented and improved prompts iteratively, and the final prompt is presented in Fig. 2 . The overall performance of the ChatGPT (gpt-3.5-turbo-16k model) is evaluated in the TCGA dataset ( n = 774), and the results are summarized in Table 1 . The accuracy of primary tumor features (pT), regional lymph node involvement (pN), overall tumor stage, and histological diagnosis are 0.87, 0.91, 0.76, and 0.99, respectively. The average accuracy of all attributes is 0.89. The coverage rates for pT, pN, overall stage and histological diagnosis are 0.97, 0.94, 0.94 and 0.96, respectively. Further details of the accuracy evaluation, F1, Kappa, recall, and precision for each attribute are summarized as confusion matrices in Fig. 3 .

Final prompt for information extraction and estimation from pathology reports.
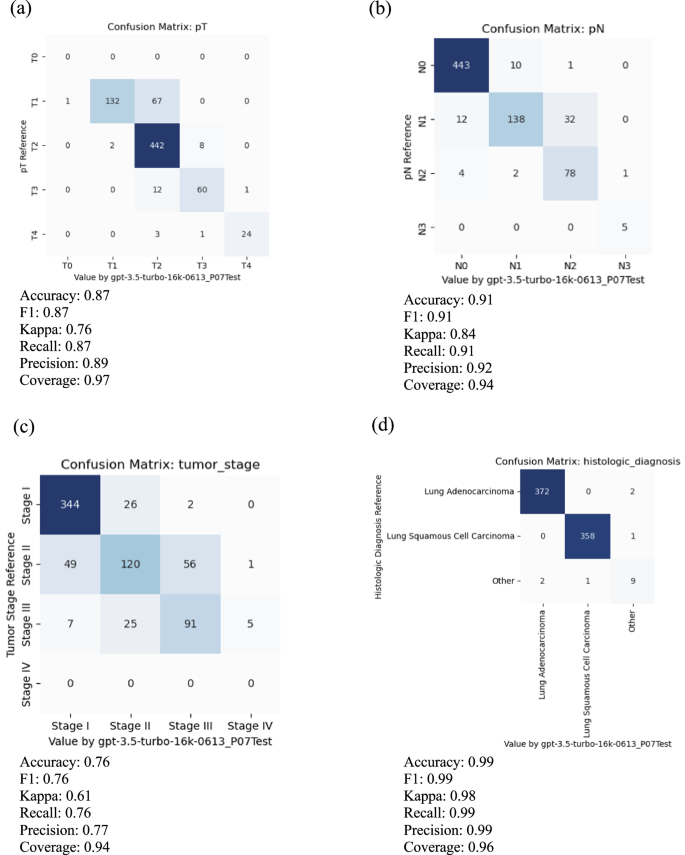
For meaningful evaluation, the cases with uncertain values, such as “Not Available”, “Not Specified”, “Cannot be determined”, “Unknown”, et al. in reference and prediction have been removed. a Primary tumor features (pT), b regional lymph node involvement (pN), c overall tumor stage, and d histological diagnosis.
Inference and Interpretation
To understand how ChatGPT reads and makes inferences from pathology reports, we demonstrated a case study using a typical pathology report in this cohort (TCGA-98-A53A) in Fig. 4a . The left panel shows part of the original pathology report, and the right panel shows the ChatGPT output with estimated pT, pN, overall stage, and histology diagnosis. For each estimate, ChatGPT gives the confidence level and the corresponding evidence it used for the estimation. In this case, ChatGPT correctly extracted information related to tumor size, tumor features, lymph node involvement, and histology information and used the AJCC staging guidelines to estimate tumor stage correctly. In addition, the confidence level, evidence interpretation, and case summary align well with the report and pathologists’ evaluations. For example, the evidence for the pT category was described as “The pathology report states that the tumor is > 3 cm and < 5 cm in greatest dimension, surrounded by lung or visceral pleura.” The evidence for tumor stage was described as “Based on the estimated pT category (T2a) and pN category (N0), the tumor stage is determined to be Stage IB according to AJCC7 criteria.” It shows that ChatGPT extracted relevant information from the note and correctly inferred the pT category based on the AJCC guideline (Supplementary Fig. 1 ) and the extracted information.
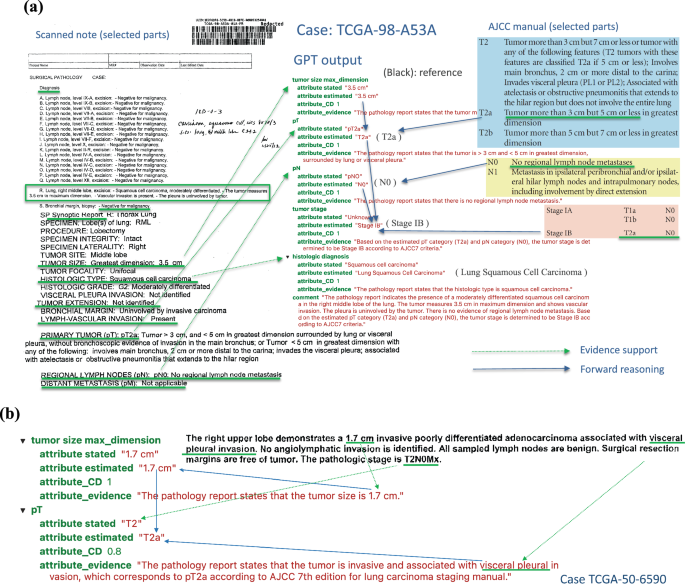
a TCGA-98-A53A. An example of a scanned pathological report (left panel) and ChatGPT output and interpretation (right panel). All estimations and support evidence are consistent with the pathologist’s evaluations. b The GPT model correctly inferred pT as T2a based on the tumor’s size and involvement according to AJCC guidelines.
In another more complex case, TCGA-50-6590 (Fig. 4b ), ChatGPT correctly inferred pT as T2a based on both the tumor’s size and location according to AJCC guidelines. Case TCGA-44-2656 demonstrates a more challenging scenario (Supplementary Fig. 2 ), where the report only contains some factual data without specifying pT, pN, and tumor stage. However, ChatGPT was able to infer the correct classifications based on the reported facts and provide proper supporting evidence.
Error analysis
To understand the types and potential reasons for misclassifications, we performed a detailed error analysis by looking into individual attributes and cases where ChatGPT made mistakes, the results of which are summarized below.
Primary tumor feature (pT) classification
In total, 768 cases with valid reports and reference values in the testing data were used to evaluate the classification performance of pT. Among them, 15 cases were reported with unknown or empty output by ChatGPT, making the coverage rate 0.97. For the remaining 753 cases, 12.6% of pT was misclassified. Among these misclassification cases, the majority were T1 misclassified as T2 (67 out of 753 or 8.9%) or T3 misclassified as T2 (12 out of 753, or 1.6%).
In most cases, ChatGPT extracted the correct tumor size information but used an incorrect rule to distinguish pT categories. For example, in the case TCGA-22-4609 (Fig. 5a ), ChatGPT stated, “Based on the tumor size of 2.0 cm, it falls within the range of T2 category according to AJCC 7th edition for lung carcinoma staging manual.” However, according to the AJCC 7 th edition staging guidelines for lung cancer, if the tumor is more than 2 cm but less than 3 cm in greatest dimension and does not invade nearby structures, pT should be classified as T1b. Therefore, ChatGPT correctly extracted the maximum tumor dimension of 2 cm but incorrectly interpreted this as meeting the criteria for classification as T2. Similarly, for case TCGA-85-A4JB, ChatGPT incorrectly claimed, “Based on the tumor size of 10 cm, the estimated pT category is T2 according to AJCC 7th edition for lung carcinoma staging manual.” According to the AJCC 7 th edition staging guidelines, a tumor more than 7 cm in greatest dimension should be classified as T3.

a TCGA-22-4609 illustrates a typical case where the GPT model uses a false rule, which is incorrect by AJCC guideline. b Case TCGA-39-5028 shows a complex case where there exist two tumors and the GPT model only capture one of them. c Case TCGA-39-5016 reveals a case where the GPT model made a mistake for getting confused with domain terminology.
Another challenging situation arose when multiple tumor nodules were identified within the lung. In the case of TCGA-39-5028 (Fig. 5b ), two separate tumor nodules were identified: one in the right upper lobe measuring 2.1 cm in greatest dimension and one in the right lower lobe measuring 6.6 cm in greatest dimension. According to the AJCC 7 th edition guidelines, the presence of separate tumor nodules in a different ipsilateral lobe results in a classification of T4. However, ChatGPT classified this case as T2a, stating, “The pathology report states the tumor’s greatest diameter as 2.1 cm”. This classification would be appropriated if the right upper lobe nodule were a single isolated tumor. However, ChatGPT failed to consider the presence of the second, larger nodule in the right lower lobe when determining the pT classification.
Regional lymph node involvement (pN)
The classification performance of pN was evaluated using 753 cases with valid reports and reference values in the testing data. Among them, 27 cases were reported with unknown or empty output by ChatGPT, making the coverage rate 0.94. For the remaining 726 cases, 8.5% of pN was misclassified. Most of these misclassification cases were N1 misclassified as N2 (32 cases). The AJCC 7th edition staging guidelines use the anatomic locations of positive lymph nodes to determine N1 vs. N2. However, most of the misclassification cases were caused by ChatGPT interpreting the number of positive nodes rather than the locations of the positive nodes. One such example is the case TCGA-85-6798. The report states, “Lymph nodes: 2/16 positive for metastasis (Hilar 2/16)”. Positive hilar lymph nodes correspond to N1 classification according to AJCC 7th edition guidelines. However, ChatGPT misclassifies this case as N2, stating, “The pathology report states that 2 out of 16 lymph nodes are positive for metastasis. Based on this information, the pN category can be estimated as N2 according to AJCC 7th edition for lung carcinoma staging manual.” This interpretation is incorrect, as the number of positive lymph nodes is not part of the criteria used to determine pN status according to AJCC 7th edition guidelines. The model misinterpreted pN2 predictions in 22 cases due to similar false assertions.
In some cases, the ChatGPT model made classification mistakes by misunderstanding the locations’ terminology. Figure 5c shows a case (TCGA-39-5016) where the ChatGPT model recognized that “6/9 peribronchial lymph nodes involved, “ corresponding with classification as N1, but ChatGPT misclassified this case as N2. By AJCC 7th edition guidelines, N2 is defined as “Metastasis in ipsilateral mediastinal and/or subcarinal lymph node(s)”. The ChatGPT model did not fully understand that terminology and made misclassifications.
Pathology tumor stage
The overall tumor stage classification performance was evaluated using 744 cases with valid reports and reference values as stage I, II and III in the testing data. Among them, 18 cases were reported as unknown or empty output by ChatGPT making the coverage rate as 0.94. For the remaining 726 cases, 23.6% of the overall stage was misclassified. Since the overall stage depends on individual pT and pN stages, the mistakes could come from misclassification of pT or pN (error propagation) or applying incorrect inference rules to determine the overall stage from pT and pN (incorrect rules). Looking into the 56 cases where ChatGPT misclassified stage II as stage III, 22 cases were due to error propagation, and 34 were due to incorrect rules. Figure 6a shows an example of error propagation (TCGA-MP-A4TK). ChatGPT misclassified the pT stage from T2a to T3, and then this mistake led to the incorrect classification of stage IIA to stage IIIA. Figure 6b illustrates a case (TCGA-49-4505) where ChatGPT made correct estimation of pT and pN but made false prediction about tumor stage by using a false rule. Among the 34 cases affected by incorrect rules, ChatGPT mistakenly inferred tumor stage as stage III for 26 cases where pT is T3 and pN is N0, respectively. For example, for case TCGA-55-7994, ChatGPT provided the evidence as “Based on the estimated pT category (T3) and pN category (N0), the tumor stage is determined to be Stage IIIA according to AJCC7 criteria”. According to AJCC7, tumors with T3 and N0 should be classified as stage IIB. Similarly, error analysis for other tumor stages shows that misclassifications come from both error propagation and applying false rules.

a Case TCGA-MP-A4TK: An example of typical errors GPT made in the experiments, i.e. GPT took false rule and further led to faulty propagation. b Case TCGA-49-4505: The GPT model made false estimation of Stage IIIA with a false rule, although it made correct inference with T2b and N1.

Histological diagnosis
The classification performance of histology diagnosis was evaluated using 762 cases with valid reports and reference values in the testing data. Among them, 17 cases were reported as either unknown or empty output by ChatGPT, making the coverage rate 0.96. For the remaining 745 cases, 6 ( < 1%) of histology types were misclassified. Among the mistakes that ChatGPT made for histology diagnosis, ChatGPT misclassified 3 of them as “other” type and 3 cases of actual “other” type (neither adenocarcinomas nor squamous cell carcinomas) as 2 adenocarcinomas and 1 squamous cell carcinoma. In TCGA-22-5485, two tumors exist: one squamous cell carcinoma and another adenocarcinoma, which should be classified as the ‘other’ type. However, ChatGPT only identified and extracted information for one tumor. In the case TCGA-33-AASB, which is the “other” type of histology, ChatGPT captured the key information and gave it as evidence: “The pathology report states the histologic diagnosis as infiltrating poorly differentiated non-small cell carcinoma with both squamous and glandular features”. However, it mistakenly estimated this case as “adenocarcinoma”. In another case (TCGA-86-8668) of adenocarcinoma, ChatGPT again captured key information and stated as evidence, “The pathology report states the histologic diagnosis as Bronchiolo-alveolar carcinoma, mucinous” but could not tell it is a subtype of adenocarcinoma. Both cases reveal that ChatGPT still has limitations in the specific domain knowledge in lung cancer pathology and the capability of correcting understanding its terminology.
Analyzing irregularities
The initial model evaluation and prompt-response review uncovered irregular scenarios: the original pathology reports may be blank, poorly scanned, or simply missing report forms. We reviewed how ChatGPT responded to these anomalies. First, when a report was blank, the prompt contained only the instruction part. ChatGPT failed to recognize this situation in most cases and inappropriately generated a fabricated case. Our experiments showed that, with the temperature set at 0 for blank reports, ChatGPT converged to a consistent, hallucinated response. Second, for nearly blank reports with a few random characters and poorly scanned reports, ChatGPT consistently converged to the same response with increased variance as noise increased. In some cases, ChatGPT responded appropriately to all required attributes but with unknown values for missing information. Last, among the 15 missing report forms in a small dataset, ChatGPT responded “unknown” as expected in only 5 cases, with the remaining 10 still converging to the hallucinated response.
Reproducibility evaluation
Since ChatGPT models (even with the same version) evolve over time, it is important to evaluate the stability and reproducibility of ChatGPT. For this purpose, we conducted experiments with the same model (“gpt-3.5-turbo-0301”), the same data, prompt, and settings (e.g., temperature = 0) twice in early April and the middle of May of 2023. The rate of equivalence between ChatGPT estimations in April and May on key attributes of interest (pT, pN, tumor stage, and histological diagnosis) is 0.913. The mean absolute error between certainty degrees in the two experiments is 0.051. Considering the evolutionary nature of ChatGPT models, we regard an output difference to a certain extent as reasonable and the overall ChatGPT 3.5 model as stable.
Comparison with other NLP methods
In order to have a clear perspective on how ChatGPT’s performance stands relative to established methods, we conducted a comparative analysis of the results generated by ChatGPT with two established methods: a keyword search algorithm and a deep learning-based Named Entity Recognition (NER) method.
Data selection and annotation
Since the keyword search and NER methods do not support zero-shot learning and require human annotations on the entity level, we carefully annotated our dataset for these traditional NLP methods. We used the same training and testing datasets as in the prompt engineering for ChatGPT. The training dataset underwent meticulous annotation by experienced medical professionals, adhering to the AJCC7 standards. This annotation process involved identifying and highlighting all relevant entities and text spans related to stage, histology, pN, and pT attributes. The detailed annotation process for the 78 cases required a few weeks of full-time work from medical professionals.
Keyword search algorithm using wordpiece tokenizer
For the keyword search algorithm, we employed the WordPiece tokenizer to segment words into subwords. We compiled an annotated entity dictionary from the training dataset. To assess the performance of this method, we calculated span similarities between the extracted spans in the validation and testing datasets and the entries in the dictionary.
Named Entity Recognition (NER) classification algorithm
For the NER classification algorithm, we designed a multi-label span classification model. This model utilized the pre-trained Bio_ClinicalBERT as its backbone. To adapt it for multi-label classification, we introduced an additional linear layer. The model underwent fine-tuning for 1000 epochs using the stochastic gradient descent (SGD) optimizer. The model exhibiting the highest overall F1 score on the validation dataset was selected as the final model for further evaluation in the testing dataset.
Performance evaluation
We evaluated the performance of both the keyword search and NER methods on the testing dataset. We summarized the predicted entities/spans and their corresponding labels. In cases where multiple related entities were identified for a specific category, we selected the most severe entities as the final prediction. Moreover, we inferred the stage information for corpora lacking explicit staging information by aggregating details from pN, pT, and diagnosis, aligning with the AJCC7 protocol. The overall predictions for stage, diagnosis, pN, and pT were compared against the ground truth table to gauge the accuracy and effectiveness of our methods. The results (Supplementary Table S1 ) show that the ChatGPT outperforms WordPiece tokenizer and NER Classifier. The average accuracy for ChatGPT, WordPiece tokenizer, and NER Classifier are 0.89, 0.51, and 0.76, respectively.
Prompt engineering process and results
Prompt design is a heuristic search process with many elements to consider, thus having a significantly large design space. We conducted many experiments to explore better prompts. Here, we share a few typical prompts and the performance of these prompts in the training data set to demonstrate our prompt engineering process.
Output format
The most straightforward prompt without special design would be: “read the pathology report and answer what are pT, pN, tumor stage, and histological diagnosis”. However, this simple prompt would make ChatGPT produce unstructured answers varying in format, terminology, and granularity across the large number of pathology reports. For example, ChatGPT may output pT as “T2” or “pT2NOMx”, and it outputs histological diagnosis as “Multifocal invasive moderately differentiated non-keratinizing squamous cell carcinoma”. The free-text answers will require a significant human workload to clean and process the output from ChatGPT. To solve this problem, we used a multiple choice answer format to force ChatGPT to pick standardized values for some attributes. For example, for pT, ChatGPT could only provide the following outputs: “T0, Tis, T1, T1a, T1b, T2, T2a, T2b, T3, T4, TX, Unknown”. For the histologic diagnosis, ChatGPT could provide output in one of these categories: Lung Adenocarcinoma, Lung Squamous Cell Carcinoma, Other, Unknown. In addition, we added the instruction, “Please make sure to output the whole set of answers together as a single JSON file, and don’t output anything beyond the required JSON file,” to emphasize the requirement for the output format. These requests in the prompt make the downstream analysis of ChatGPT output much more efficient. In order to know the certainty degree of ChatGPT’s estimate and the evidence, we asked ChatGPT to provide the following 4 outputs for each attribute/variable: extracted value as stated in the pathology report, estimated value based on AJCC 7th edition for lung carcinoma staging manual, the certainty degree of the estimation, and the supporting evidence for the estimation. The classification accuracy of this prompt with multiple choice output format (prompt v1) in our training data could achieve 0.854.
Evidence-based inference
One of the major concerns for LLM is that the results from the model are not supported by any evidence, especially when there is not enough information for specific questions. In order to reduce this problem, we emphasize the use of evidence for inference in the prompt by adding this instruction to ChatGPT: “Please ensure to make valid inferences for attribute estimation based on evidence. If there is no available evidence provided to make an estimation, please answer the value as “Unknown.” In addition, we asked ChatGPT to “Include “comment” as the last key of the JSON file.” After adding these two instructions (prompt v2), the performance of the classification in the training data increased to 0.865.
Chain of thought prompting by asking intermediate questions
Although tumor size is not a primary interest for diagnosis and clinical research, it plays a critical role in classifying the pT stage. We hypothesize that if ChatGPT pays closer attention to tumor size, it will have better classification performance. Therefore, we added an instruction in the prompt (prompt v3) to ask ChatGPT to estimate: “tumor size max_dimension: [<the greatest dimension of tumor in Centimeters (cm)>, ‘Unknown’]” as one of the attributes. After this modification, the performance of the classification in the training data increased to 0.90.
Providing examples
Providing examples is an effective way for humans to learn, and it should have similar effects for ChatGPT. We provided a specific example to infer the overall stage based on pT and pN by adding this instruction: “Please estimate the tumor stage category based on your estimated pT category and pN category and use AJCC7 criteria. For example, if pT is estimated as T2a and pN as N0, without information showing distant metastasis, then by AJCC7 criteria, the tumor stage is “Stage IB”.” After this modification (prompt v4), the performance of the classification in the training data increased to 0.936.
Although we can further refine and improve prompts, we decided to use prompt v4 as the final model and apply it to the testing data and get the final classification accuracy of 0.89 in the testing data.
ChatGPT-4 performance
LLM evolves rapidly and OpenAI just released the newest GPT-4 Turbo model (GPT-4-1106-preview) in November 2023. To compare this new model with GPT-3.5-Turbo, we applied this newest GPT model GPT-4-1106 to analyze all the lung cancer pathology notes in the testing data. The classification result and the comparison with the GPT-3.5-Turbo-16k are summarized in Supplementary Table 1 . The results show that GPT-4-turbo performs better in almost every aspect; overall, the GPT-4-turbo model increases performance by over 5%. However, GPT-4-Turbo is much more expensive than GPT-3.5-Turbo. The performance of GPT-3.5-Turbo-16k is still comparable and acceptable. As such, this study mainly focuses on assessing GPT-3.5-Turbo-16k, but highlights the fast development and promise of using LLM to extract structured data from clinical notes.
Analyzing osteosarcoma data
To demonstrate the broader application of this method beyond lung cancer, we collected and analyzed clinical notes from pediatric osteosarcoma patients. Osteosarcoma, the most common type of bone cancer in children and adolescents, has seen no substantial improvement in patient outcomes for the past few decades 18 . Histology grades and margin status are among the most important prognostic factors for osteosarcoma. We collected pathology reports from 191 osteosarcoma cases (approved by UTSW IRB #STU 012018-061). Out of these, 148 cases had histology grade information, and 81 had margin status information; these cases were used to evaluate the performance of the GPT-3.5-Turbo-16K model and our prompt engineering strategy. Final diagnoses on grade and margin were manually reviewed and curated by human experts, and these diagnoses were used to assess ChatGPT’s performance. All notes were de-identified prior to analysis. We applied the same prompt engineering strategy to extract grade and margin information from these osteosarcoma pathology reports. This analysis was conducted on our institution’s private Azure OpenAI platform, using the GPT-3.5-Turbo-16K model (version 0613), the same model used for lung cancer cases. ChatGPT accurately classified both grades (with a 98.6% accuracy rate) and margin status (100% accuracy), as shown in Supplementary Fig. 3 . In addition, Supplementary Fig. 4 details a specific case, illustrating how ChatGPT identifies grades and margin status from osteosarcoma pathology reports.
Since ChatGPT’s release in November 2022, it has spurred many potential innovative applications in healthcare 19 , 20 , 21 , 22 , 23 . To our knowledge, this is among the first reports of an end-to-end data science workflow for prompt engineering, using, and rigorously evaluating ChatGPT in its capacity of batch-processing information extraction tasks on large-scale clinical report data.
The main obstacle to developing traditional medical NLP algorithms is the limited availability of annotated data and the costs for new human annotations. To overcome these hurdles, particularly in integrating problem-specific information and domain knowledge with LLMs’ task-agnostic general knowledge, Augmented Language Models (ALMs) 24 , which incorporate reasoning and external tools for interaction with the environment, are emerging. Research shows that in-context learning (most influentially, few-shot prompting) can complement LLMs with task-specific knowledge to perform downstream tasks effectively 24 , 25 . In-context learning is an approach of training through instruction or light tutorial with a few examples (so called few-shot prompting; well instruction without any example is called 0-shot prompting) rather than fine-tuning or computing-intensive training, which adjusts model weights. This approach has become a dominant method for using LLMs in real-world problem-solving 24 , 25 , 26 . The advent of ALMs promises to revolutionize almost every aspect of human society, including the medical and healthcare domains, altering how we live, work, and communicate. Our study shows the feasibility of using ChatGPT to extract data from free text without extensive task-specific human annotation and model training.
In medical data extraction, our study has demonstrated the advantages of adopting ChatGPT over traditional methods in terms of cost-effectiveness and efficiency. Traditional approaches often require labor-intensive annotation processes that may take weeks and months from medical professionals, while ChatGPT models can be fine-tuned for data extraction within days, significantly reducing the time investment required for implementation. Moreover, our economic analysis revealed the cost savings associated with using ChatGPT, with processing over 900 pathology reports incurring a minimal monetary cost (less than $10 using GPT 3.5 Turbo and less than $30 using GPT-4 Turbo). This finding underscores the potential benefits of incorporating ChatGPT into medical data extraction workflows, not only for its time efficiency but also for its cost-effectiveness, making it a compelling option for medical institutions and researchers seeking to streamline their data extraction processes without compromising accuracy or quality.
A critical requirement for effectively utilizing an LLM is crafting a high-quality “prompt” to instruct the LLM, which has led to the emergence of an important methodology referred to as “prompt engineering.” Two fundamental principles guide this process: firstly, the provision of appropriate context, and secondly, delivering clear instructions about subtasks and the requirements for the desired response and how it should be presented. For a single query for one-time use, the user can experiment with and revise the prompt within the conversation session until a satisfactory answer is obtained. However, prompt design can become more complex when handling repetitive tasks over many input data files using the OpenAI API. In these instances, a prompt must be designed according to a given data feed while maintaining the generality and coverage for various input data features. In this study, we found that providing clear guidance on the output format, emphasizing evidence-based inference, providing chain of thought prompting by asking for tumor size information, and providing specific examples are critical in improving the efficiency and accuracy of extracting structured data from the free-text pathology reports. The approach employed in this study effectively leverages the OpenAI API for batch queries of ChatGPT services across a large set of tasks with similar input data structures, including but not limited to pathology reports and EHR.
Our evaluation results show that the ChatGPT (gpt-3.5-turbo-16k) achieved an overall average accuracy of 89% in extracting and estimating lung cancer staging information and histology subtypes compared to pathologist-curated data. This performance is very promising because some scanned pathology reports included in this study contained random characters, missing parts, typos, varied formats, and divergent information sections. ChatGPT also outperformed traditional NLP methods. Our case analysis shows that most misclassifications were due to a lack of knowledge of detailed pathology terminology or very specialized information in the current versions of ChatGPT models, which could be avoided with future model training or fine-tuning with more domain-specific knowledge.
While our experiments reveal ChatGPT’s strengths, they also underscore its limitations and potential risks, the most significant being the occasional “hallucination” phenomenon 27 , 28 , where the generated content is not faithful to the provided source content. For example, the responses to blank or near-blank reports reflect this issue, though these instances can be detected and corrected due to convergence towards an “attractor”.
The phenomenon of ‘hallucination’ in LLMs presents a significant challenge in the field. It is important to consider several key factors to effectively address the challenges and risks associated with ChatGPT’s application in medicine. Since the output of an LLM depends on both the model and the prompt, mitigating hallucination can be achieved through improvements in GPT models and prompting strategies. From a model perspective, model architecture, robust training, and fine-tuning on a diverse and comprehensive medical dataset, emphasizing accurate labeling and classification, can reduce misclassifications. Additionally, enhancing LLMs’ comprehension of medical terminology and guidelines by incorporating feedback from healthcare professionals during training and through Reinforcement Learning from Human Feedback (RLHF) can further diminish hallucinations. Regarding prompt engineering strategies, a crucial method is to prompt the GPT model with a ‘chain of thought’ and request an explanation with the evidence used in the reasoning. Further improvements could include explicitly requesting evidence from input data (e.g., the pathology report) and inference rules (e.g., AJCC rules). Prompting GPT models to respond with ‘Unknown’ when information is insufficient for making assertions, providing relevant context in the prompt, or using ‘embedding’ of relevant text to narrow down the semantic subspace can also be effective. Harnessing hallucination is an ongoing challenge in AI research, with various methods being explored 5 , 27 . For example, a recent study proposed “SelfCheckGPT” approach to fact-check black-box models 29 . Developing real-time error detection mechanisms is crucial for enhancing the reliability and trustworthiness of AI models. More research is needed to evaluate the extent, impacts, and potential solutions of using LLMs in clinical research and care.
When considering using ChatGPT and similar LLMs in healthcare, it’s important to thoughtfully consider the privacy implications. The sensitivity of medical data, governed by rigorous regulations like HIPAA, naturally raises concerns when integrating technologies like LLMs. Although it is a less concern to analyze public available de-identified data, like the lung cancer pathology notes used in this study, careful considerations are needed for secured healthcare data. More secured OpenAI services are offered by OpenAI security portal, claimed to be compliant to multiple regulation standards, and Microsoft Azure OpenAI, claimed could be used in a HIPAA-compliant manner. For example, de-identified Osteosarcoma pathology notes were analyzed by Microsoft Azure OpenAI covered by the Business Associate Agreement in this study. In addition, exploring options such as private versions of these APIs, or even developing LLMs within a secure healthcare IT environment, might offer good alternatives. Moreover, implementing strong data anonymization protocols and conducting regular security checks could further protect patient information. As we navigate these advancements, it’s crucial to continuously reassess and adapt appropriate privacy strategies, ensuring that the integration of AI into healthcare is both beneficial and responsible.
Despite these challenges, this study demonstrates our effective methodology in “prompt engineering”. It presents a general framework for using ChatGPT’s API in batch queries to process large volumes of pathology reports for structured information extraction and estimation. The application of ChatGPT in interpreting clinical notes holds substantial promise in transforming how healthcare professionals and patients utilize these crucial documents. By generating concise, accurate, and comprehensible summaries, ChatGPT could significantly enhance the effectiveness and efficiency of extracting structured information from unstructured clinical texts, ultimately leading to more efficient clinical research and improved patient care.
In conclusion, ChatGPT and other LLMs are powerful tools, not just for pathology report processing but also for the broader digital transformation of healthcare documents. These models can catalyze the utilization of the rich historical archives of medical practice, thereby creating robust resources for future research.
Data processing, workflow, and prompt engineering
The lung cancer data we used for this study are publicly accessible via CDSA ( https://cancer.digitalslidearchive.org/ ) and TCGA ( https://cBioPortal.org ), and they are de-identified data. The institutional review board at the University of Texas Southwestern Medical Center has approved this study where patient consent was waived for using retrospective, de-identified electronic health record data.
We aimed to leverage ChatGPT to extract and estimate structured data from these notes. Figure 7a displays our process. First, scanned pathology reports in PDF format were downloaded from TCGA and CDSA databases. Second, R package pdftools, an optical character recognition tool, was employed to convert scanned PDF files into text format. After this conversion, we identified reports with near-empty content, poor scanning quality, or missing report forms, and those cases were excluded from the study. Third, the OpenAI API was used to analyze the text data and extract structured data elements based on specific prompts. In addition, we extracted case identifiers and metadata items from the TCGA metadata file, which was used to evaluate the model performance.

a Illustration of the use of OpenAI API for batch queries of ChatGPT service, applied to a substantial volume of clinical notes — pathology reports in our study. b A general framework for integrating ChatGPT into real-world applications.
In this study, we implemented a problem-solving framework rooted in data science workflow and systems engineering principles, as depicted in Fig. 7b . An important step is the spiral approach 30 to ‘prompt engineering’, which involves experimenting with subtasks, different phrasings, contexts, format specifications, and example outputs to improve the quality and relevance of the model’s responses. It was an iterative process to achieve the desired results. For the prompt engineering, we first define the objective: to extract information on TNM staging and histology type as structured attributes from the unstructured pathology reports. Second, we assigned specific tasks to ChatGPT, including estimating the targeted attributes, evaluating certainty levels, identifying key evidence of each attribute estimation, and generating a summary as output. The output was compiled into a JSON file. In this process, clinicians were actively formulating questions and evaluating the results.
Our study used the “gpt-3.5-turbo” model, accessible via the OpenAI API. The model incorporates 175 billion parameters and was trained on various public and authorized documents, demonstrating specific Artificial General Intelligence (AGI) capabilities 5 . Each of our queries sent to ChatGPT service is a “text completion” 31 , which can be implemented as a single round chat completion. All LLMs have limited context windows, constraining the input length of a query. Therefore, lengthy pathology reports combined with the prompt and ChatGPT’s response might exceed this limit. We used OpenAI’s “tiktoken” Python library to estimate the token count to ensure compliance. This constraint has been largely relaxed by the newly released GPT models with much larger context windows. We illustrate the pseudocode for batch ChatGPT queries on a large pathology report set in Supplementary Fig. 5 .
Model evaluation
We evaluated the performance of ChatGPT by comparing its output with expert-curated data elements provided in the TCGA structured data using the testing data set. Some staging records in the TCGA structured data needed to be updated; our physicians curated and updated those records. To mimic a real-world setting, we processed all reports regardless of data quality to collect model responses. For performance evaluation, we only used valid reports providing meaningful text and excluded the reports with near-empty content, poor scanning quality, and missing report forms, which were reported as irregular cases. We assessed the classification accuracy, F1, Kappa, recall, and precision for each attribute of interest, including pT, pN, overall stage, and histology types, and presented results as accuracy and confusion matrices. Missing data were excluded from the accuracy evaluation, and the coverage rate was reported for predicted values as ‘unknown’ or empty output.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The lung cancer dataset we used for this study is “Pan-Lung Cancer (TCGA, Nat Genet2016)”, ( https://www.cbioportal.org/study/summary?id=nsclc_tcga_broad_2016 ) and the “luad” and “lusc” subsets from CDSA ( https://cancer.digitalslidearchive.org/ ). We have provided a reference regarding how to access the data 32 . We utilized the provided APIs to retrieve clinical information and pathology reports for the LUAD (lung adenocarcinoma) and LUSC (lung squamous cell carcinoma) cohorts. The pediatric data are the EHR data from UTSW clinic services. The data is available from the corresponding author upon reasonable request and IRB approval.
Code availability
All codes used in this paper were developed using APIs from OpenAI. The prompt for the API is available in Fig. 2 . Method-specific code is available from the corresponding author upon request.
Vaswani, A. et al. Attention is all you need. Adv. Neural Info. Processing Syst. 30 , (2017).
Devlin, J. et al. Bert: Pre-training of deep bidirectional transformers for language understanding . arXiv preprint arXiv:1810.04805, 2018.
Radford, A. et al. Improving language understanding by generative pre-training . OpenAI: https://cdn.openai.com/research-covers/language-unsupervised/language_understanding_paper.pdf (2018).
Touvron, H. et al. LLaMA: Open and efficient foundation language models . arXiv preprint arXiv:2302.13971 (2023).
OpenAi, GPT-4 Technical Report . arXiv:2303.08774: https://arxiv.org/pdf/2303.08774.pdf (2023).
Anil, R. et al. Palm 2 technical report . arXiv preprint arXiv:2305.10403 (2023).
Turner, B. E. W. Epic, Microsoft bring GPT-4 to EHRs .
Landi, H. Microsoft’s Nuance integrates OpenAI’s GPT-4 into voice-enabled medical scribe software .
Hao, T. et al. Health Natural Language Processing: Methodology Development and Applications. JMIR Med Inf. 9 , e23898 (2021).
Article Google Scholar
Pathak, J., Kho, A. N. & Denny, J. C. Electronic health records-driven phenotyping: challenges, recent advances, and perspectives. J. Am. Med. Inform. Assoc. 20 , e206–e211 (2013).
Article PubMed PubMed Central Google Scholar
Crichton, G. et al. A neural network multi-task learning approach to biomedical named entity recognition. BMC Bioinforma. 18 , 368 (2017).
Wang, J. et al. Document-Level Biomedical Relation Extraction Using Graph Convolutional Network and Multihead Attention: Algorithm Development and Validation. JMIR Med Inf. 8 , e17638 (2020).
Liu, Y. et al. Roberta: A robustly optimized BERT pretraining approach . arXiv preprint arXiv:1907.11692 (2019).
Rasmy, L. et al. Med-BERT: pretrained contextualized embeddings on large-scale structured electronic health records for disease prediction. npj Digit. Med. 4 , 86 (2021).
Wu, H. et al. A survey on clinical natural language processing in the United Kingdom from 2007 to 2022. npj Digit. Med. 5 , 186 (2022).
Amin, M. B. et al. AJCC cancer staging manual . 1024: Springer 2017.
Goldstraw, P. et al. The IASLC Lung Cancer Staging Project: Proposals for the Revision of the TNM Stage Groupings in the Forthcoming (Seventh) Edition of the TNM Classification of Malignant Tumours. J. Thorac. Oncol. 2 , 706–714 (2007).
Article PubMed Google Scholar
Yang, D. M. et al. Osteosarcoma Explorer: A Data Commons With Clinical, Genomic, Protein, and Tissue Imaging Data for Osteosarcoma Research. JCO Clin. Cancer Inform. 7 , e2300104 (2023).
The Lancet Digital, H., ChatGPT: friend or foe? Lancet Digital Health . 5 , e102 (2023).
Nature, Will ChatGPT transform healthcare? Nat. Med. 29 , 505–506 (2023).
Patel, S. B. & Lam, K. ChatGPT: the future of discharge summaries? Lancet Digit. Health 5 , e107–e108 (2023).
Article CAS PubMed Google Scholar
Ali, S. R. et al. Using ChatGPT to write patient clinic letters. Lancet Digit. Health 5 , e179–e181 (2023).
Howard, A., Hope, W. & Gerada, A. ChatGPT and antimicrobial advice: the end of the consulting infection doctor? Lancet Infect. Dis. 23 , 405–406 (2023).
Mialon, G. et al. Augmented language models: a survey . arXiv preprint arXiv:2302.07842 (2023).
Brown, T. et al. Language Models are Few-Shot Learners . Curran Associates, Inc. (2020).
Wei, J. et al. Chain of thought prompting elicits reasoning in large language models . Adv Neural Inf Processing Syst 35 , 24824–24837 (2022).
Ji, Z. et al. Survey of Hallucination in Natural Language Generation. ACM Comput. Surv. 55 , 1–38 (2023).
Alkaissi, H. & S. I. McFarlane, Artificial Hallucinations in ChatGPT: Implications in Scientific Writing. Cureus , (2023).
Manakul, P. A. Liusie, & M. J. F. Gales, SelfCheckGPT: Zero-Resource Black-Box Hallucination Detection for Generative Large Language Models . 2023.
Boehm, B. W. A spiral model of software development and enhancement. Computer 21 , 61–72 (1988).
OpenAi. OpenAI API Documentation . Available from: https://platform.openai.com/docs/guides/text-generation .
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6 , 1–19 (2013).
Download references
Acknowledgements
This work was partially supported by the National Institutes of Health [P50CA70907, R35GM136375, R01GM140012, R01GM141519, R01DE030656, U01CA249245, and U01AI169298], and the Cancer Prevention and Research Institute of Texas [RP230330 and RP180805].
Author information
Authors and affiliations.
Quantitative Biomedical Research Center, Peter O’Donnell School of Public Health, University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd, Dallas, TX, USA 75390, USA
Jingwei Huang, Donghan M. Yang, Ruichen Rong, Kuroush Nezafati, Colin Treager, Shidan Wang, Xian Cheng, Yujia Guo, Guanghua Xiao, Xiaowei Zhan & Yang Xie
Department of Pathology, University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd, Dallas, TX, USA 75390, USA
Department of Pediatrics, University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd, Dallas, TX, USA 75390, USA
Laura J. Klesse
Department of Internal Medicine, University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd, Dallas, TX, USA 75390, USA
Eric D. Peterson
You can also search for this author in PubMed Google Scholar
Contributions
J.H., Y.X., X.Z. and G.X. designed the study. X.Z., K.N., C.T. and J.H. prepared, labeled, and curated lung cancer datasets. D.M.Y., X.C., Y.G., L.J.K. prepared, labeled, and curated osteosarcoma datasets. Z.C. provided critical inputs as pathologists. Y.X., G.X., E.P. provided critical inputs for the study. J.H. implemented experiments with ChatGPT. R.R. and K.N. implemented experiments with N.L.P. J.H., Y.X., G.X. and S.W. conducted data analysis. Y.X., G.X., J.H., X.Z., D.M.Y. and R.R. wrote the manuscript. All co-authors read and commented on the manuscript.
Corresponding authors
Correspondence to Xiaowei Zhan or Yang Xie .
Ethics declarations
Competing interests.
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplemental figures and tables, reporting summary, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Huang, J., Yang, D.M., Rong, R. et al. A critical assessment of using ChatGPT for extracting structured data from clinical notes. npj Digit. Med. 7 , 106 (2024). https://doi.org/10.1038/s41746-024-01079-8
Download citation
Received : 24 July 2023
Accepted : 14 March 2024
Published : 01 May 2024
DOI : https://doi.org/10.1038/s41746-024-01079-8
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.
- Open access
- Published: 29 April 2024
Identification of Novel NSD1 variations in four Pediatric cases with sotos Syndrome
- Zhuo Ren 1 na1 ,
- Ling Yue 2 na1 ,
- Hua-ying Hu 3 na1 ,
- Xiao-lin Hou 4 ,
- Wen-qi Chen 4 ,
- Zhe Dong 1 &
- Jing Zhang 4
BMC Medical Genomics volume 17 , Article number: 116 ( 2024 ) Cite this article
Metrics details
Sotos syndrome (SOTOS) is an uncommon genetic condition that manifests itself with the following distinctive features: prenatal overgrowth, facial abnormalities, and intellectual disability. This disorder is often associated with haploinsufficiency of the nuclear receptor-binding SET domain protein 1 ( NSD1 )gene. We investigated four pediatric cases characterized by early-onset overgrowth and developmental delay. The primary objective of this study was to achieve accurate genetic diagnoses.
Design&Methods
A sequential analysis approach comprising chromosomal karyotyping, whole exome sequencing, and microarray analysis was conducted.
All four cases exhibited variations in the NSD1 gene, with the identification of four previously unreported de novo variants, each specific to one case.Specifically, Case 1 carried the NSD1 (NM_022455): c.2686 C > T(p.Q896X) variant, Case 2 had the NSD1 (NM_022455): c.2858_2859delCT(p.S953X) variant, Case 3 displayed a chromosomal aberration, chr5: 5q35.2q35.3(176,516,604–176,639,249)×1, which encompassed the 5′-untranslated region of NSD1 , and Case 4 harbored the NSD1 (NM_022455): c.6397T > G(p.C2133G) variant.
This study not only provided precise diagnoses for these cases but also supplied significant evidence to facilitate informed consultations. Furthermore, our findings expanded the spectrum of mutations associated with SOTOS.
Peer Review reports
Introduction
Sotos syndrome (SOTOS), previously known as cerebral gigantism (OMIM #117,550), was first described by Juan Fernandez Sotos et al. in 1964 [ 1 ]. This neurologic disease is characterized by excessive growth from the prenatal stage to childhood, accompanied with distinctive facial abnormalities (large skull, pointed chin, and acromegaly), advanced bone age, seizures, occasional brain abnormalities, and intellectual disability [ 2 , 3 ]. The estimated prevalence of SOTOS is 1 in 14,000 live births. The prominent features include behavioral abnormalities, often within the autistic spectrum, abnormal cranial magnetic resonance imaging (MRI)/computed tomography (CT) findings, cardiac anomalies, joint hyperlaxity with or without pes planus, neonatal complications, maternal preeclampsia, scoliosis, renal anomalies, and seizures [ 2 ]. However, the definitive clinical diagnostic criteria of SOTOS has been challenging because its characteristic features may be potentially misdiagnosed for other genetic disorders, such as Weaver–Smith syndrome (OMIM #277,590) [ 4 ], Malan syndrome (OMIM #614,753) [ 5 ], Cohen–Gibson syndrome (OMIM #617,561) [ 6 ], and other similar disorders [ 7 ]. Thus, genetic techniques are considered crucial for the accurate differential diagnosis of these conditions.
SOTOS follows an autosomal dominant inheritance pattern with complete penetrance, and > 95% of the affected individuals carry a de novo pathogenic variant [ 2 ]. The nuclear receptor-binding SET domain protein 1 ( NSD1 ) gene (OMIM *606,681), located on chromosome 5q35.3 and containing 23 exons, is the only gene that has been identified to cause SOTOS [ 8 ]. The diagnosis of SOTOS in the proband can be achieved by identifying either a heterozygous pathogenic (or potentially pathogenic) variant in NSD1 or a deletion encompassing NSD1 through molecular genetic testing [ 3 , 9 , 10 ]. So far,>500 disease-causing variants have been documented and indexed in the Human Gene Mutation Database ( https://www.hgmd.cf.ac.uk/ ; 20,230,801). These variants encompass a diverse spectrum of mutations, including missense, truncating, partial gene deletions, splice site alterations, and 5q35 microdeletions, and all of them lead to NSD1 haploinsufficiency. The past two decades have seen significant advancements in genetic technology with notable enhancements in the detection efficiency and classification accuracy of SOTOS-related variations [ 11 ]. While the genotype–phenotype correlation of SOTOS has been explored in a few studies [ 3 , 9 , 12 , 13 , 14 ], the consensus is that individuals carrying the 5q35 microdeletion have milder overgrowth but more severe intellectual disabilities than those with the intragenic NSD1 pathogenic variant [ 2 ]. However, several other symptoms lack well-established associations with specific variants, necessitating further investigation.
The present study presents clinical and genetic findings, as well as imaging data of four pediatric cases diagnosed with SOTOS. The identification of the novel NSD1 variants in these cases not only contributes to a more comprehensive understanding of the condition but also expands the mutation spectrum associated with SOTOS.
Materials and methods
This study obtained approval from the Ethics Committee of Shijiazhuang Obstetrics and Gynecology Hospital (approval no. 20,230,160). Informed consent was acquired from the legal guardians of all minor participants. All procedures conducted in this study adhered to the principles outlined in the Declaration of Helsinki of 1964, along with its subsequent revisions and pertinent ethical standards.
Participants and clinical assessment
Four cases of suspected SOTOS fetuses were admitted to our center between December 2020 and January 2023. A comprehensive clinical assessment comprising general clinic monitoring in the outpatient department and an MRI examination, followed by a thorough investigation of the family history, was conducted.
Karyotyping and copy number variation analyses
A comprehensive genetic diagnosis was performed with peripheral blood (PB) samples collected from the probands for traditional chromosomal karyotyping using G-binding for total chromosomal anomalies detection [ 15 ].
Genomic DNA extraction was performed with the PB samples collected from the probands and parents using the QIAamp DNA Midi Kit (Qiagen, Germany). Chromosomal microarray analysis (CMA) was conducted using the CytoScan 750 K SNP-array (Affymetrix Inc., USA) following manufacturer’s protocols to investigate clinically significant genomic copy number variations (CNVs).
- Whole exome sequencing
Sequence variants were detected in the proband samples using whole exome sequencing (WES), in accordance with our previously described method [ 16 ].Briefly, Agilent Sure Select Human Exon Sequence Capture Kit (Agilent, USA) was used for the enrichment of target region sequences. Quantitative polymerase chain reaction (qPCR) and Agilent Bioanalyzer 2100 (Agilent, USA) were utilized to assess the DNA library quality by determining the size, content, and distribution. Subsequently, the NovaSeq6000 platform (Illumina) was used to sequence DNA samples having ? ∼ 150 bp paired-end reads, with approximately 300 pM per sample using the NovaSeq Reagent kit. The sequenced raw reads were aligned to the human reference genome (accession no.: hg19/GRCh37) using the Burrows Wheeler Aligner tool(quality level Q30% > 90% and the quality criteria are listed at https://www.illumina.com/science/technology/next-generation-sequencing/plan-experiments/quality-scores.html).Picard(v1.57 ) Variant calling was employed for the purpose of removing PCR duplicates using the Genome Analysis Tool Kit ( https://software.broadinstitute.org/gatk/ ) and the Verita Trekker® Variants Detection system (v2.0; Berry Genomics, China). This process was performed using the ANNOVAR (v2.0) and Enliven® Variants Annotation Interpretation systems (Berry Genomics [ 17 ], and the guidelines recommended by the American College of Medical Genetics and Genomics(ACMG) were followed [ 18 ]. For variant pathogenicity interpretation, three frequency databases (ExAC_EAS, http://exac.broadinstitute.org ; gnomAD_exome_EAS, http://gnomad.broadinstitute.org ; 1000G_2015aug_eas, https://www.internationalgenome.org ), and Human Gene Mutation Database (pro v2019) were used as reference. Additionally, the REVEL (rare exome variant ensemble learner; an integrated pathogenicity prediction approach) [ 19 ] and the pLI (probability of being loss-of-function intolerant; indicating tolerance of truncating variants) scores were used.
The sequence verification was performed with Sanger sequencing using the 3500DX Genetic Analyzer (Applied Biosystems, Thermo Fisher Scientific, USA). Quantitative fluorescence PCR (qfPCR) was employed to confirm the CNV results (detailed method described in Supplementary Material 1 ).
Conservatism and structural analyses
The evolutionary conservation of affected amino acid residues related to the respective missense variant was analyzed using MEGA 7 ( http://www.megasoftware.net/previousVersions.php ) with default parameters. Structure models for the wildtype and missense variant were constructed using the AF-Q96L73-F1 structure prediction ( https://alphafold.ebi.ac.uk/entry/Q96L73 ) with the Swiss-Model program ( https://swissmodel.expasy.org/)includin g default parameters.
Clinical presentation
The four cases featured in this study underwent initial assessment before reaching the age of 2 years, with the earliest case observed at just 4 months of age. Case 1, who underwent the initial evaluation at 1 year and 11 months, presented with impaired language and cognitive development and displayed unsteady walking. This individual was delivered via cesarean section at full term and encountered oxygen deprivation during birth. The physical examination unveiled several notable characteristics, including a head circumference measuring 50 cm (approximately at the 95th centile), a distinctive facial appearance marked by a wide interocular distance and prominent forehead, above-average height, and lags in response. The patient’s posture exhibited atypical characteristics, including hip flexion, strephexopodia, and a tendency to experience falls. Fine motor skills of both hands were notably restricted, with limited finger pinch flexibility. His responsiveness to commands was inadequate, and he encountered challenges in feature recognition. Cardiac ultrasound examination unveiled the presence of an atrial septal defect, while cerebral MRI revealed delayed myelination, bilateral ventricular enlargement, and dysplasia of the corpus callosum (Fig. 1 A-D).
Case, a four-month-old male infant, presented at our outpatient clinic with complaints of head instability and difficulty in turning over. During the physical examination, it was noted that he had a head circumference measuring 47 cm (above the 99th centile), wide interocular distance, and ears set lower than usual. Furthermore, he displayed delayed responses to both sounds and objects. His Albert Motor Scale score was 35 (equivalent to the 6th month, at the 10th–25th centile, indicating low-to-medium motor development). Cardiac ultrasound examination revealed a loose and thickened left ventricular apex myocardial tissue structure, along with mild regurgitation at the bicuspid and tricuspid valves. Cerebral MRI findings in Case 2 indicated delayed myelination, substantial enlargement of the bifrontal extracerebral space and bilateral ventricles, as well as thinning of the corpus callosum, collectively suggestive of brain dysplasia(Figs. 1 E–M).
Moving on to Case 3, a 5-month-old female infant, she was also referred to our outpatient clinic due to head instability and difficulty in turning over. Her physical examination revealed a head circumference measuring 47.5 cm (above the 99th centile), wide-set eyes, a pointed chin, ears set lower than typical, and a high-arched palate. Additionally, she exhibited delayed responses to sounds and objects and had elevated muscle tone in her lower limbs. Cardiac ultrasound revealed the presence of an atrial septal defect and an increased forward velocity of the pulmonary valve. Cerebral MRI demonstrated delayed myelination, a small subdural hemorrhage in the right parietal lobe, widening of the extracerebral space in the bilateral frontotemporal region, irregular enlargement of the bilateral ventricles, and a slightly thinner corpus callosum(Figs. 1 N–Q).
Case, a 5-month-old girl, visited our clinic for delayed motor development. Physical examination revealed almost similar manifestations as those observed in Case 3 (head circumference of 47.5 cm, wide-set eyes, pointed chin, low-set ears, and high-arched palate). Cardiac ultrasound revealed an atrial septal defect and increased forward velocity of the pulmonary valve. Cerebral MRI demonstrated delayed myelination, a small amount of subdural hemorrhage in the right parietal lobe, widening of the bilateral frontotemporal extracerebral space, irregular widening of the bilateral ventricles, and slightly thinner callosum (Fig. 1 R–V).
Genetic assessment
The pedigree diagram for each case is shown in Fig. 2 . Karyotyping analysis results were normal for all four cases. In Case 3, CMA detected a de novo heterozygous microdeletion, chr5: 5q35.2q35.3(176,516,604–176,639,249)×1, encompassing the 5′-UTR fragment of NSD1, which was subsequently confirmed using qfPCR (Fig. 2 C). No abnormalities in the CMA results were observed in the remaining three cases.
WES successfully identified the causative variants in the remaining three cases, and the results were validated with Sanger sequencing. Specifically, Case 1 carried a de novo truncating variant, NSD1 (NM_022455): c.2686 C > T(p.Q896X) (Fig. 2 A); Case 2 harbored a de novo frame-shift variant, NSD1 (NM_022455): c.2858_2859delCT(p.S953X), resulting in early translation termination (Fig. 2 B); and Case 4 exhibited a de novo missense variant, NSD1 (NM_022455): c.6397T > G(p.C2133G)(Fig. 2 D). All the three sequence variations identified were novel and have not been previously reported.
The details of the identified variations in the four cases are presented in Table 1 .
Missense variant analyses
We also identified a solitary missense variant, NSD1 (NM_022455): c.6397T > G(p.C2133G). According to MEGA 7 results, the p.C2133 residue was conserved across multiple species (Fig. 3 A). Structural modeling analysis revealed that p.C2133G primarily affected local hydrogen bond formation. In the wildtype model, the C2133 site formed hydrogen bonds with K2140, R2117, and H2162 within the β-helix. The interactions of C2133 with K2140 and H2162 occurred inside the β-helix, and the interactions between C2133 and R2117 occurred outside the β-helix (Fig. 3 B, upper right). In the missense variant model, C2133G exhibited hydrogen bonds with K2140 and R2117,but it was absent between G2133 and H2162. Additionally, the distance of the hydrogen bonds formed between G2133 and K2140 as well as G2133 and R2117 was altered (Fig. 3 B, lower right).
Neonatal overgrowth is a common clinical concern with a multifaceted etiology. It can result from various factors, including maternal lifestyle choices during pregnancy, gestational diabetes, neonatal hyperinsulinism, or congenital syndromes [ 20 ]. Overgrowth syndromes encompass a diverse array of rare conditions characterized by either segmental or generalized overgrowth, often accompanied by additional features such as macrocephaly, visceromegaly, and other symptoms [ 7 , 21 ]. Recent advancements in molecular technology, particularly next-generation sequencing (NGS), have significantly improved the efficiency of identifying novel causative genes and have increased the detection rate for overgrowth syndromes.
SOTOS represents a distinctive form of overgrowth primarily involving nervous system development. Many patients with SOTOS exhibit varying degrees of learning disabilities, ranging from mild to severe, along with behavioral abnormalities and symptoms related to autism spectrum disorder [ 22 ]. More than 40% of SOTOS cases are accompanied with seizures [ 3 ]. Skeletal manifestations in this disorder are noted, such as scoliosis(50%), along with flat feet and genu valgum or genu varum, suggesting a possible association with hyperlaxity [ 2 ]. Although cranial MRI/CT is not the definitive diagnostic criterion, majority of patients with SOTOS exhibit characteristic findings, such as ventricular dilation (particularly in the trigone region), midline anomalies (agenesis or hypoplasia of the corpus callosum, cavum septum pellucidum, and mega cisterna magna), small cerebellar vermis, and cerebral atrophy [ 23 ].
NSD1 , the only gene known to cause SOTOS, encodes the histone methyltransferase protein (H3K36 methylation), which is involved in gene transcription control and as an epigenetic marker [ 24 ]. NSD1 expression has been detected in numerous tissues across diverse organisms. NSD1 expression levels are upregulated in the pancreas, brain, hematopoietic organs (lymphoid tissues and bone marrow), and the male reproductive tract. NSD1 gene cloning revealed that NSD1 expression is evident in multiple tissues, and western blotting assays revealed two transcript variants, attributable to the alternative splicing of exons 3 and 4 [ 25 ]. Mutations in several other genes with similar functionality, such as SETD2, DNMT3A, or APC2, can also lead to SOTOS-like syndromes [ 26 , 27 , 28 ].
All four cases in our study exhibited clinical and radiographic findings consistent with previous reports, but a definitive diagnosis was highly dependent on genetic techniques. Molecular genetic methods successfully identified the diagnostic variants in the four cases examined in our study, encompassing three sequence variants and one intragenic CNV. To the best of our knowledge, all four variations have not been previously reported. Haploinsufficiency as the pathogenic mechanism of NSD1 is well established [ 8 ]. Case 1 carried a de novo truncating variant, NSD1 (NM_022455): c.2686 C > T(p.Q896X); Case 2 harbored a de novo frame-shift variant, NSD1 (NM_022455): c.2858_2859delCT(p.S953X), resulting in early translation termination; Case 3 possessed a de novo heterozygous microdeletion, chr5: 5q35.2q35.3(176,516,604–176,639,249)×1, as detected via CMA and encompassing the 5′-UTR fragment of NSD1 . The variants in Cases 1–3 are null variants that either hinder proper gene expression or yield truncated proteins prone to degradation. Thus, the variants in Cases 1–3 are pathogenic. The missense variant, NSD1 (NM_022455): c.6397T > G(p.C2133G), in Case 4 resides in the plant homeodomain (PHD) 5 domain of the NSD1 protein, exhibiting considerable conservation across species. PHD acts as a C4HC3 zinc finger-like motif within nuclear proteins that are involved in chromatin modulation and epigenetics [ 29 ]. This variant significantly influenced local hydrogen bond formation, potentially compromising the stability of the protein’s three-dimensional structure. Based on these findings and the alignment with the ACMG’s interpretation criteria [ 18 ], we designated this variant as “likely pathogenic.” Recent reports demonstrated that NSD1 mutations result in dysregulated DNA methylation and transcription of bivalent developmental genes in SOTOS [ 30 , 31 ]. Consequently, transcriptome sequencing and other epigenetic methods are promising tools for the detection of neurodysplasia, especially in patients with negative genomic results [ 32 ].
Genetically, all four cases exhibited NSD1 -associated variations, with the identification of four novel de novo variants specific to each case. The parents of the four cases were wildtype, and thus, future pregnancy for these couples is accompanied with a very low risk of SOTOS recurrence. However, targeted prenatal diagnosis should be recommended for these couples in subsequent pregnancies.
In summary, this study presents a definitive genetic diagnosis for four children exhibiting overgrowth and central nervous system abnormalities. The genetic variants revealed in this study expand the mutation spectrum of NSD1 associated with SOTOS. Additionally, this research underscores the potential of NGS technology in facilitating the differential diagnosis of neuropathic conditions characterized by nonspecific clinical phenotypes.
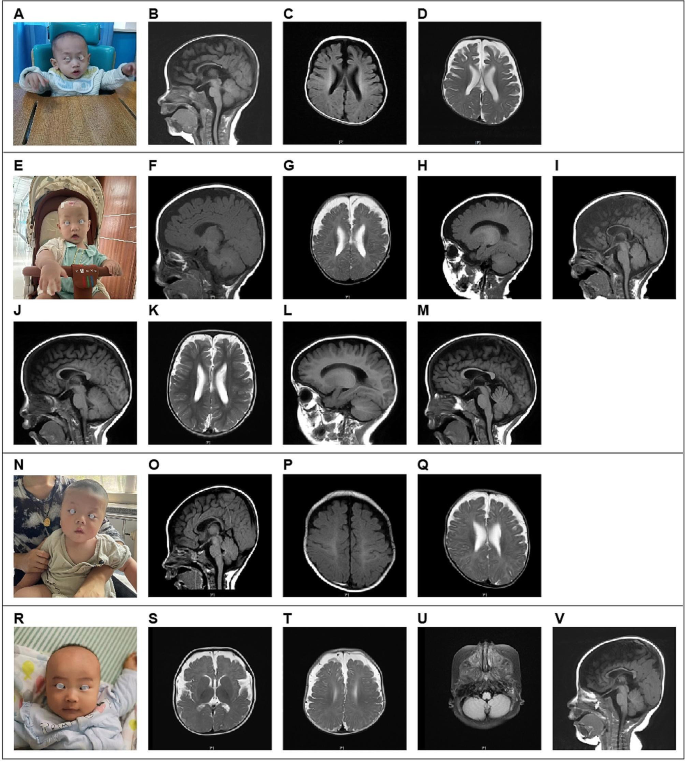
MRI indications in cerebral magnetic resonance imaging for the four cases. A – D : Case 1 had delayed myelination, widening of the bilateral ventricles, and dysplasia of the corpus callosum. E – I : (4 months and 9 days): Case 2 possessed a smaller brain mass, exhibited widening of the bilateral ventricles, and incomplete closure of the transparent compartments; the cerebral sulcus was slightly wider, the bifrontotemporal extracerebral space was significantly wider, and the corpus callosum was narrow; and a long T2 signal occurred in the bilateral mastoid; J – M : (1 year and 4 months): In Case 2, a reduction in the widening of the bifrontotemporal extraencephalic space and an increased in the widening of the bilateral ventricles was noted. N – Q : Case 3 had slightly delayed myelination, widening of the bifrontotemporal extracerebral space, irregular widening of the bilateral ventricles, and slightly thinner corpus callosum. R – V : Case 4 had a smaller frontal lobe volume, an old bleeding lesion in the left occipital lobe, a softened lesion with hemosiderosis, low signal in the right cerebellar hemisphere, significant widening of the bilateral frontotemporal extracerebral space, and thin corpus callosum
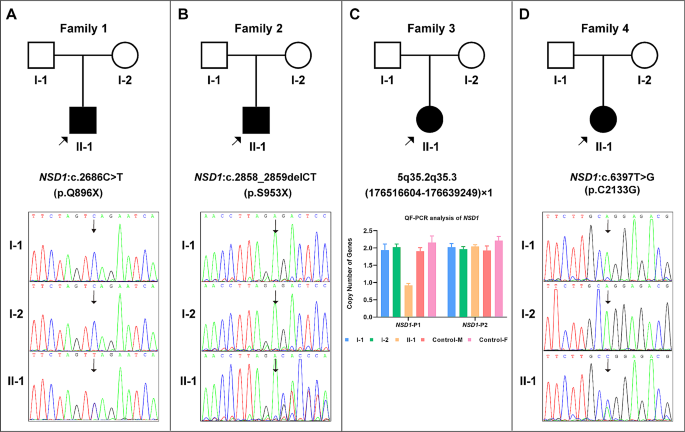
Genetic findings for the four probands. A : Case 1, de novo NSD1 (NM_022455): c.2686 C > T(p.Q896X) variant. B :Case 2, de novo NSD1 (NM_022455): c.2858_2859delCT(p.S953X) variant. C : Case 3, de novo heterozygous microdeletion, chr5: 5q35.2q35.3(176,516,604–176,639,249)×1 ( NSD1 -P1), containing the 5′-UTR region of NSD1 ; the result on the right side ( NSD1 -P2, chr5:177,235,826–177,235,945) suggests that the coding area of NSD1 was normal. D : Case 4, de novo NSD1 (NM_022455): c.6397T > G(p.C2133G) variant
Analysis of the missense variant. A : The C2133 residue is conserved across species. B : NSD1: p.C2133G variation significantly affected hydrogen bond formation at this location
Data availability
The datasets presented in this study can be found in online repositories. The sequencing results have been deposited to the Figshare repository and can be accessed via the following links: https://doi.org/10.6084/m9.figshare.24135969 .
Sotos JF, Dodge PR, Muirhead D, Crawford JD, Talbot NB. CEREBRAL GIGANTISM IN CHILDHOOD. A SYNDROME OF EXCESSIVELY RAPID GROWTH AND ACROMEGALIC FEATURES AND A NONPROGRESSIVE NEUROLOGIC DISORDER. N Engl J Med. 1964;271:109–16.
Article CAS PubMed Google Scholar
Tatton-Brown K, Cole TR, Rahman N, Sotos Syndrome. 2004 Dec 17 [Updated 2022 Dec 1]. In: Adam MP, Mirzaa GM, Pagon RA, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2023. https://www.ncbi.nlm.nih.gov/books/NBK1479/ .
Tatton-Brown K, Douglas J, Coleman K, Baujat G, Cole TR, Das S, Horn D, Hughes HE, Temple IK, Faravelli F, et al. Genotype-phenotype associations in Sotos syndrome: an analysis of 266 individuals with NSD1 aberrations. Am J Hum Genet. 2005;77(2):193–204.
Article CAS PubMed PubMed Central Google Scholar
Gibson WT, Hood RL, Zhan SH, Bulman DE, Fejes AP, Moore R, Mungall AJ, Eydoux P, Babul-Hirji R, An J, et al. Mutations in EZH2 cause Weaver syndrome. Am J Hum Genet. 2012;90(1):110–8.
Priolo M, Schanze D, Tatton-Brown K, Mulder PA, Tenorio J, Kooblall K, Acero IH, Alkuraya FS, Arias P, Bernardini L, et al. Further delineation of Malan syndrome. Hum Mutat. 2018;39(9):1226–37.
Cohen AS, Tuysuz B, Shen Y, Bhalla SK, Jones SJ, Gibson WT. A novel mutation in EED associated with overgrowth. J Hum Genet. 2015;60(6):339–42.
Edmondson AC, Kalish JM. Overgrowth syndromes. J Pediatr Genet. 2015;4(3):136–43.
Article PubMed PubMed Central Google Scholar
Kurotaki N, Imaizumi K, Harada N, Masuno M, Kondoh T, Nagai T, Ohashi H, Naritomi K, Tsukahara M, Makita Y, et al. Haploinsufficiency of NSD1 causes Sotos syndrome. Nat Genet. 2002;30(4):365–6.
Kurotaki N, Harada N, Shimokawa O, Miyake N, Kawame H, Uetake K, Makita Y, Kondoh T, Ogata T, Hasegawa T, et al. Fifty microdeletions among 112 cases of Sotos syndrome: low copy repeats possibly mediate the common deletion. Hum Mutat. 2003;22(5):378–87.
Ferilli M, Ciolfi A, Pedace L, Niceta M, Radio FC, Pizzi S, Miele E, Cappelletti C, Mancini C, Galluccio T et al. Genome-Wide DNA Methylation Profiling Solves Uncertainty in Classifying NSD1 Variants. Genes (Basel) 2022, 13(11).
Testa B, Conteduca G, Grasso M, Cecconi M, Lantieri F, Baldo C, Arado A, Andraghetti L, Malacarne M, Milani D et al. Molecular analysis and reclassification of NSD1 gene variants in a cohort of patients with clinical suspicion of Sotos Syndrome. Genes (Basel) 2023, 14(2).
Ha K, Anand P, Lee JA, Jones JR, Kim CA, Bertola DR, Labonne JD, Layman LC, Wenzel W, Kim HG. Steric clash in the SET domain of histone methyltransferase NSD1 as a cause of Sotos Syndrome and its genetic heterogeneity in a Brazilian cohort. Genes (Basel) 2016, 7(11).
Foster A, Zachariou A, Loveday C, Ashraf T, Blair E, Clayton-Smith J, Dorkins H, Fryer A, Gener B, Goudie D, et al. The phenotype of Sotos syndrome in adulthood: a review of 44 individuals. Am J Med Genet C Semin Med Genet. 2019;181(4):502–8.
Article PubMed Google Scholar
Conteduca G, Testa B, Baldo C, Arado A, Malacarne M, Candiano G, Garbarino A, Coviello DA, Cantoni C. Identification of alternative transcripts of NSD1 gene in Sotos Syndrome patients and healthy subjects. Gene. 2023;851:146970.
Arsham MS, Barch MJ, Lawce HJ. The AGT cytogenetics laboratory manual. New Jersey: John Wiley & Sons Inc.; 2017.
Book Google Scholar
Yang K, Liu Y, Wu J, Zhang J, Hu HY, Yan YS, Chen WQ, Yang SF, Sun LJ, Sun YQ, et al. Prenatal cases reflect the complexity of the COL1A1/2 Associated Osteogenesis Imperfecta. Genes (Basel). 2022;13(9):1578.
Wang KLM, Hakonarson H. ANNOVAR: functional annotation of genetic variants from next-generation sequencing data. NUCLEIC ACIDS RES. 2010;38:e164.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. GENET MED. 2015;17(5):405–24.
Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, Musolf A, Li Q, Holzinger E, Karyadi D, et al. REVEL: an Ensemble Method for Predicting the pathogenicity of rare missense variants. AM J HUM GENET. 2016;99(4):877–85.
Manor J, Lalani SR. Overgrowth Syndromes-Evaluation, diagnosis, and management. Front Pediatr. 2020;8:574857.
Brioude F, Toutain A, Giabicani E, Cottereau E, Cormier-Daire V, Netchine I. Overgrowth syndromes - clinical and molecular aspects and tumour risk. Nat Rev Endocrinol. 2019;15(5):299–311.
Lane C, Milne E, Freeth M. Characteristics of Autism Spectrum Disorder in Sotos Syndrome. J Autism Dev Disord. 2017;47(1):135–43.
Waggoner DJ, Raca G, Welch K, Dempsey M, Anderes E, Ostrovnaya I, Alkhateeb A, Kamimura J, Matsumoto N, Schaeffer GB, et al. NSD1 analysis for Sotos syndrome: insights and perspectives from the clinical laboratory. Genet Med. 2005;7(8):524–33.
Martin-Herranz DE, Aref-Eshghi E, Bonder MJ, Stubbs TM, Choufani S, Weksberg R, Stegle O, Sadikovic B, Reik W, Thornton JM. Screening for genes that accelerate the epigenetic aging clock in humans reveals a role for the H3K36 methyltransferase NSD1. Genome Biol. 2019;20(1):146.
Kurotaki N, Harada N, Yoshiura K, Sugano S, Niikawa N, Matsumoto N. Molecular characterization of NSD1, a human homologue of the mouse Nsd1 gene. Gene. 2001;279(2):197–204.
Luscan A, Laurendeau I, Malan V, Francannet C, Odent S, Giuliano F, Lacombe D, Touraine R, Vidaud M, Pasmant E, et al. Mutations in SETD2 cause a novel overgrowth condition. J Med Genet. 2014;51(8):512–7.
Almuriekhi M, Shintani T, Fahiminiya S, Fujikawa A, Kuboyama K, Takeuchi Y, Nawaz Z, Nadaf J, Kamel H, Kitam AK, et al. Loss-of-function mutation in APC2 causes sotos Syndrome features. Cell Rep. 2015;10(9):1585–98.
Tlemsani C, Luscan A, Leulliot N, Bieth E, Afenjar A, Baujat G, Doco-Fenzy M, Goldenberg A, Lacombe D, Lambert L, et al. SETD2 and DNMT3A screen in the sotos-like syndrome French cohort. J Med Genet. 2016;53(11):743–51.
Romero VI, Arias-Almeida B, Aguiar SA. NSD1 gene evolves under episodic selection within primates and mutations of specific exons in humans cause Sotos syndrome. BMC Genomics. 2022;23(1):849.
Brennan K, Zheng H, Fahrner JA, Shin JH, Gentles AJ, Schaefer B, Sunwoo JB, Bernstein JA, Gevaert O. NSD1 mutations deregulate transcription and DNA methylation of bivalent developmental genes in Sotos syndrome. Hum Mol Genet. 2022;31(13):2164–84.
Hamagami N, Wu DY, Clemens AW, Nettles SA, Li A, Gabel HW. NSD1 deposits histone H3 lysine 36 dimethylation to pattern non-CG DNA methylation in neurons. Mol Cell. 2023;83(9):1412–e14281417.
Lecoquierre F, Quenez O, Fourneaux S, Coutant S, Vezain M, Rolain M, Drouot N, Boland A, Olaso R, Meyer V, et al. High diagnostic potential of short and long read genome sequencing with transcriptome analysis in exome-negative developmental disorders. Hum Genet. 2023;142(6):773–83.
Download references
Acknowledgements
We thank the patients and their families for their participation in this study.
The work was supported by the Key Research and Development Program of Hebei Province (No.2137720D).
Author information
Zhuo Ren, Ling Yue, Hua-ying Hu equal contributors to this study.
Authors and Affiliations
Department of Obstetrics and Gynecology, Peking University International Hospital, Beijing, China
Zhuo Ren, Ya Tan & Zhe Dong
Department of Pediatric Neurology Rehabilitation, Hebei Children’s Hospital, Shijiazhuang, Hebei, China
Birth Defects Prevention and Control Technology Research Center, Medical Innovation Research Division of Chinese PLA General Hospital, Beijing, China
Hua-ying Hu
Prenatal Diagnosis Center, Hebei Key Laboratory of Maternal and Fetal Medicine, Shijiazhuang Key Laboratory of Reproductive Health, Shijiazhuang Obstetrics and Gynecology Hospital, 16 Tangu-North Street, Shijiazhuang, Hebei, China
Xiao-lin Hou, Wen-qi Chen & Jing Zhang
You can also search for this author in PubMed Google Scholar
Contributions
JZ and ZR, conception and design; LY, WC and JZ, resources; ZR, HH, and YT, methodology; XH and ZD, data analysis. ZR was responsible for first manuscript drafting, and our authors approved these submissions.
Corresponding author
Correspondence to Jing Zhang .
Ethics declarations
Ethics approval and consent to participate.
This study received approval from the Ethics Committee of Shijiazhuang Obstetrics and Gynecology Hospital (approval no. 20230160). Informed consent was taken from guardian’s of all minor participants. All procedures in this study were conducted in accordance with the principles outlined in the Declaration of Helsinki 1964, as well as its subsequent amendments and relevant ethical standards.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1
Rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Ren, Z., Yue, L., Hu, Hy. et al. Identification of Novel NSD1 variations in four Pediatric cases with sotos Syndrome. BMC Med Genomics 17 , 116 (2024). https://doi.org/10.1186/s12920-024-01889-5
Download citation
Received : 22 August 2023
Accepted : 24 April 2024
Published : 29 April 2024
DOI : https://doi.org/10.1186/s12920-024-01889-5
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- SOTOS syndrome
- Structural analyses
BMC Medical Genomics
ISSN: 1755-8794
- Submission enquiries: [email protected]
- General enquiries: [email protected]
Disclaimer » Advertising
- HealthyChildren.org

- Previous Article
- Next Article
Bottom Line
Improving pediatric osteomyelitis evaluation with a risk score.
- Split-Screen
- Article contents
- Figures & tables
- Supplementary Data
- Peer Review
- CME Quiz Close Quiz
- Open the PDF for in another window
- Get Permissions
- Cite Icon Cite
- Search Site
Improving Pediatric Osteomyelitis Evaluation With a Risk Score. AAP Grand Rounds May 2024; 51 (5): 56. https://doi.org/10.1542/gr.51-5-56
Download citation file:
- Ris (Zotero)
- Reference Manager
Source: Stephan AM , Platt S , Levine DA , et al . A novel risk score to guide the evaluation of acute hematogenous osteomyelitis in children . Pediatrics . 2024 ; 153 ( 2 ): e2023063153 ; doi: https://doi.org/10.1542/peds.2023-063153 . Google Scholar
Investigators from multiple institutions conducted a case-control study to develop a clinical prediction rule to identify children being evaluated for possible musculoskeletal (MSK) infection who are at high risk for acute hematogenous osteomyelitis (AHO). For the study, data on children >90 days and <18 years old seen because of suspected MSK infection at 23 pediatric EDs between 2017 and 2019, were reviewed. Cases were eligible patients with an ICD-10 code for AHO, confirmed by medical record review. Controls were children evaluated for MSK infection, including laboratory and radiographic testing, without a diagnosis of osteomyelitis at the index ED visit or within 30 days. Each case was matched to 2 controls by study site and closest date to case ED visit. Multiple demographic, clinical, and laboratory characteristics were compared in cases and controls. Receiver operator characteristic (ROC) analysis was used to identify threshold values for continuous variables, such as laboratory results, that maximized their predictive value. Multivariate logistic regression was performed to identify independent predictors of AHO and calculate odds ratios (ORs) for individual variables. Based on these results, the investigators used a systematic process to choose items for a clinical prediction rule to classify patients as at high risk of AHO. The area under the curve (AUC), sensitivity, and specificity of the prediction rule were determined.
A total of 1,135 cases with AHO were matched with 2,270 controls. Among the cases, 98.3% presented with pain, 94.9% with impaired mobility, and 79.0% with a history of fever. The most common sites of osteomyelitis were the tibia (22.4%), femur (20.9%), pelvis (18.4%), and fibula (10.9%). A bacterial pathogen was identified in 67.4% of children with AHO. In the multivariate model, multiple characteristics were identified as predictors of AHO, including C-reactive protein (CRP) >2.0 mg/dL (OR, 4.68; 95% CI, 3.67, 5.98), erythrocyte sedimentation rate (ESR) >25 mm/hr (OR, 3.70; 95% CI, 2.95, 4.92), duration of illness >3 days (OR, 3.27; 95% CI, 2.63, 4.07), history of fever (OR, 2.60; 95% CI, 2.06, 3.28), and fever in the ED (OR, 2.98; 95% CI, 2.35, 3.77). After combining ED fever and history of fever into 1 clinical finding, these 4 characteristics were used in the clinical prediction rule, with 1 point assigned to each positive finding. The AUC of the prediction rule was 0.892. Using a score of ≥3 as the threshold value, the sensitivity of the rule in predicting a diagnosis of AHO was 0.78 (95% CI, 0.75, 0.80), and specificity was 0.86 (95% CI. 0.85, 0.88).
The authors conclude that the clinical prediction rule they derived was useful in identifying children at high risk for AHO.
Dr Loyal has disclosed no financial relationship relevant to this commentary. This commentary does not contain a discussion of an unapproved/investigative use of a commercial product/device.
Prompt recognition and initiation of treatment of uncomplicated AHO can be confounded by a variety of signs and symptoms in children of all ages. 1 Delayed diagnosis or complicated infections can involve multiple bones and/or soft tissue, and be associated with bacteremia, septic thrombophlebitis, venous thrombosis, pathologic fractures, or adverse effects on bone growth. 1 Children with AHO often present with pain and fever, and the diagnosis is further supported by elevated acute inflammatory markers, leukocytosis, MRI, and a bone biopsy. In the current study, investigators propose a 4-point risk score to guide clinical decision making for AHO in children with a MSK complaint. The authors suggest that this information can help guide clinicians considering pursuing an MRI or bone biopsy. While potentially safeguarding against unnecessary sedation for diagnostic imaging or procedures, validated clinical prediction tools also can help reduce variation in management and promote antibiotic stewardship.
A similar prediction tool in MSK infections, the Kocher criteria for septic arthritis, is widely used to distinguish septic arthritis from transient synovitis in a child with an inflamed hip. 2 As in the current study, there are 4 criteria (non-weight bearing, temperature >38.5°C, WBC >12,000/mm 3 , and ESR >40 mm/hr); in the presence of all 4, the probability for septic arthritis in the validation study was 93%. 2 Of note, some subsequent researchers have been unable to replicate the same findings, 3 and the prediction rule sometimes is applied inappropriately to other joints. 3 In the current study, following application of likelihood ratios to population estimates of AHO, patients with all 4 factors in the risk score would have an absolute risk of 37.9% to 49.1%, which is suboptimal. A challenge with accurately diagnosing AHO are the mimickers (eg, pyomyositis, septic arthritis, tumors), in which fever and elevated inflammatory markers are not unusual.
Future validation studies will be of great interest to clinicians managing suspected MSK infections.
In children being evaluated for a suspected MSK infection, a 4-point risk score can potentially inform the need to pursue further diagnostic testing for AHO. (See AAP Grand Rounds . 2023;50[6]:63.) 4
Advertising Disclaimer »
Citing articles via
Email alerts.

Affiliations
- Editorial Board
- Pediatrics On Call
- Online ISSN 1556-362X
- Print ISSN 1099-6605
- Pediatrics Open Science
- Hospital Pediatrics
- Pediatrics in Review
- AAP Grand Rounds
- Latest News
- Pediatric Care Online
- Red Book Online
- Pediatric Patient Education
- AAP Toolkits
- AAP Pediatric Coding Newsletter
First 1,000 Days Knowledge Center
Institutions/librarians, group practices, licensing/permissions, integrations, advertising.
- Privacy Statement | Accessibility Statement | Terms of Use | Support Center | Contact Us
- © Copyright American Academy of Pediatrics
This Feature Is Available To Subscribers Only
Sign In or Create an Account

IMAGES
VIDEO
COMMENTS
Fifteen months before this admission, the patient was evaluated at the pediatric clinic of another hospital. On examination, the pulse was 95 beats per minute and the blood pressure 117/78 mm Hg.
Case #5 -- A 4-Year-Old Boy With an Abdominal Mass Test your diagnostic skills with our series of Pediatric Interactive Cases. Clinical Case, May 12, 2003. Interactive Case #4 - A Child With ...
A 13-year-old boy presents to his primary care provider with a 5-day history of abdominal pain and a 2-day history of diarrhea and vomiting. He describes the quality of the abdominal pain as sharp, originating in the epigastric region and radiating to his back, and exacerbated by movement. Additionally, he has had several episodes of nonbloody, nonbilious vomiting and watery diarrhea. His ...
11-year-old boy with testicular pain and rash. William A. Frese, MD, MPH. January 19th 2024. An 11-year-old boy presented to the emergency department complaining of left testicular pain for 2 days, described as intermittent and stabbing, which ranged between 5 and 8 of 10 in intensity. Read the full case to see if you can correctly diagnose the ...
A 13-year-old boy with a recent diagnosis of ulcerative colitis presented with fever and chest pain to the emergency department. The sharp pain had begun 2 days previous and radiated to his shoulders. It was constant, although it improved with sitting up and leaning forward. On the day before, he had developed a fever to 38.3°C. His past medical history was notable for a diagnosis of ...
Nishant Banait, Sai Vamshi Varanasi, Abinash Nayak, Kushal Talukder. 21 March 2024. Endobronchial lesion in a premature neonate. Zakhar Shchomak, Lia Oliveira, Ana Saianda, Teresa Bandeira
View by: Case Topic Case Number. Accidental Ingestion of Opioids. Acute Epstein-Barr Virus (Infectious Mononucleosis) Acute Lymphoblastic Leukemia. Acute Otitis Media. Acute Postinfectious Glomerulonephritis. Adolescent Substance Use Disorder. Alopecia Areata Online Only. Anaphylactoid Purpura (Henoch-Schönlein Purpura)
We present 3 scenarios that are commonly encountered in clinical practice: isolated elevation of low-density lipoprotein cholesterol (LDL-C), combined dyslipidemia, and severe hypertriglyceridemia. Treatment with statin is indicated when the LDL-C is ≥190 mg/dL (4.9 mmol/L) in children ≥10 years of age. For LDL-C levels between 130 and 189 ...
ia, fever, nuchal rigidity, myalgias, and fatigue. Character and significance of symptoms vary by patient age. Symptoms of infection may improve spontaneously or worsen, becoming potentially lethal. Early recognition and treatment of meningitis are crucial to prevent morbidity and mortality. The case reviewed in this article focuses on viral meningitis in a pediatric patient that may be ...
08 Mar 2024. 28 Feb 2024. 26 Feb 2024. Case Reports in Pediatrics publishes case reports and case series related to pediatric subspecialities such as adolescent medicine, cardiology, critical care, dentistry, developmental and behavioral medicine, endocrinology, gastroenterology etc.
Cases in Pediatric Acute Care presents over 100 real-world pediatric acute care cases, each including a brief patient history, a detailed history of present illness, presenting signs and symptoms, vital signs, and physical examination findings. Ideal for developing a systematic approach to diagnosis, evaluation, and treatment, this resource provides students and advanced practitioners with the ...
Pediatrics is the official peer-reviewed journal of the American Academy of Pediatrics. Pediatrics publishes original research, clinical observations, and special feature articles in the field of pediatrics, as broadly defined. ... All authors must have been engaged in the advocacy work described in the case study. Additional authors can be ...
This calculation is based on 80 mLs/kg total blood volume in a 30 Kg child or 2,400 mLs. With a 25% blood loss we expected a 25% decrease in hemoglobin level, which is exactly what occurred. The preoperative hemoglobin level was 12.5 g/dL, and the postoperative level was 9.4. Because the tumor was adherent to the ventricles, the plan was to ...
Results from published studies suggest that omalizumab should be continued for > 1 year [77, 78]. In a retrospective study of adults and children with uncontrolled severe asthma treated with omalizumab, the response to treatment was 'excellent' in 52.5% of patients, particularly in the subgroup of children aged 6 to 11 years . After the ...
Case 1. Previously healthy 11-year-old male presents with respiratory distress and dehydration. On exam: Afebrile. Patient is ill appearing with sunken eyes. Appears breathless. Cannot complete full sentence without taking another breath. Taking deep and fast respirations, RR 27. Lungs clear to auscultation.
Introduction. Following are case studies of children with typical developmental behavioral issues that may require a host of referrals and recommendations.. Case Studies Case 1: Case 2: Case 3:
Abstract. Nephrotic syndrome (NS) is a glomerular disorder typically characterized by gross proteinuria, hypoalbuminemia, hyperlipidemia, and peripheral oedema. We report the case of a 2-year-old male toddler weighing 15 kg with a 1-week history of swelling around the eyes and both legs, and generalized body swelling.
J.S. is a former 32-weeks'-gestation infant who presents as a new patient for his 4-month health supervision visit. His birth history includes a 4-week neonatal intensive care unit stay caused by difficulty feeding, but ever since his discharge, he has been doing well on fortified human (breast) milk of 22 kcal/fl oz.
The causes of constipation in children are poorly understood, and for the individual child the cause is often unknown. Organic constipation is due to an underlying pathology. It occurs in approximately 5%-10% of cases and is more likely to present in the first month of life. Causes in babies and toddlers include:
This retrospective case series study was conducted from 2013 to 2023, in infants less than 42 days of age who had virologically confirmed HSV infection. We report an overall rate of neonatal herpes of 5.5 per 100,000 live births a year and an incidence of symptomatic cases of 1.2 per 100,000 live births a year.
BACKGROUND: Pediatric ARDS is associated with significant morbidity and mortality. High-quality data from clinical trials in children are limited due to numerous barriers to their design and execution. Here we describe the collaborative development of a master protocol as a tool to address some of these barriers and support the conduct of pediatric ARDS studies.
The first of these Advocacy Case Studies, by Bronstein et al., (10.1542/peds.2018-1886) focuses on how legislation was created and then passed by the California State legislature in 2016 to require the state to develop processes to comply with any new federal recommendations for newborn screening within two years of that recommendation. In this ...
In pediatric osteosarcoma dataset, ChatGPT-3.5 accurately classified both grades and margin status with accuracy of 98.6% and 100% respectively. ... we demonstrated a case study using a typical ...
Sotos syndrome (SOTOS) is an uncommon genetic condition that manifests itself with the following distinctive features: prenatal overgrowth, facial abnormalities, and intellectual disability. This disorder is often associated with haploinsufficiency of the nuclear receptor-binding SET domain protein 1 (NSD1)gene. We investigated four pediatric cases characterized by early-onset overgrowth and ...
Source: Stephan AM, Platt S, Levine DA, et al. A novel risk score to guide the evaluation of acute hematogenous osteomyelitis in children. Pediatrics. 2024;153(2):e2023063153; doi: 10.1542/peds.2023-063153.Investigators from multiple institutions conducted a case-control study to develop a clinical prediction rule to identify children being evaluated for possible musculoskeletal (MSK ...