
- school Campus Bookshelves
- menu_book Bookshelves
- perm_media Learning Objects
- login Login
- how_to_reg Request Instructor Account
- hub Instructor Commons
- Download Page (PDF)
- Download Full Book (PDF)
- Periodic Table
- Physics Constants
- Scientific Calculator
- Reference & Cite
- Tools expand_more
- Readability
selected template will load here
This action is not available.


9: Proteins - An Introduction
- Last updated
- Save as PDF
- Page ID 431831
- 9.1: Properties of Carboxylic Acids and Amines Carboxylic acids are weak acids, Therefore they are proton donors to water. Amines are weak bases. So amines are proton acceptors from water.
- 9.2: Amino Acids Amino acids can be classified based on the characteristics of their distinctive side chains as nonpolar, polar neutral, negatively charged, or positively charged.
- 9.3: The Peptide Bond A dehydration-condensation reaction forms a peptide bond between 2 amino acids. An amine group of one amino acid undergoes a reaction with the carboxylic acid group of another amino acid forming an amide functional group.
- 9.4: Oxidation of Thiols The oxidation of two thiol functional groups leads to the formation of the disulfide. Disulfide linkages (–S–S–) between protein chains are extremely important in tertiary and quaternary structure of the protein.
- 9.5: Protein Structure Proteins may be fibrous, or globular. A protein may have up to four levels of structure.
- 9.6: Denaturation of Proteins A wide variety of reagents and conditions can cause a protein to denture. Denaturation leads to loss of biological activity of the protein due to loss of three-dimensional structure.
- 9.7: Digestion of Proteins During digestion, the peptide bonds in proteins undergo hydrolysis to amino acids. The digestion process of proteins begins in the stomach and continues in the small intestine.
- 9.8: End of Chapter Problems This problem set is based on Chapter 9 topics.
Here’s a fact that will boggle your mind. There is a retinal protein named after the cartoon character Pikachu! It is called Pikachurin. In fact, proteins are known to have unique names. Proteins are complex biomolecules that are made up of smaller units known as amino acids . Let us learn about them in detail.
Suggested Videos:

Structure of Proteins
Due to different rearrangement of amino acids, the structure of proteins divides into four types:
- Primary- the covalent linkages of the proteins

(Source: Wikipedia)
- Secondary- the linear peptide chains fold either into an alpha-helical structure(coiled) or a beta-pleated structure(sheets) which contain hydrogen bonds .

- Tertiary- The arrangement and interconnection of proteins into specific loops and bends forms the tertiary structures. This structure contains hydrogen, ionic and disulfide bonds.
- Quarternary- this structure is proteins containing more than one peptide chain.
Proteins are made up of smaller units known as amino acids and the bond linking them is known as a peptide bond. This bond is formed when the carboxyl group (-COOH) of one amino acid bonds with the amino group (– NH 2 ) of another amino acid releasing a molecule of water (H2O). A peptide may be dipeptide, tripeptide, and polypeptide.

Classification of Proteins
Classification of proteins is done on the basis of the following:
- Constitution
- Nature of molecules
On the basis of shape
- Fibrous protein(Scleroprotein): We can find these proteins in animals and are insoluble in water. Fibrous proteins are resistant to proteolytic enzymes and are coiled and exist in threadlike structures to form fibres. e.g. collagen, actin, and myosin, keratin in hair, claws, feathers, etc.
- Globular proteins: These proteins, unlike fibrous proteins are soluble in water. They are made up of polypeptides that are coiled about themselves to form oval or spherical molecules e.g. albumin, insulin, and hormones like oxytocin, etc.
On the basis of Constitution
- Simple proteins: These proteins are made up of amino acids only. e.g. albumins, globulins, prolamins, etc.
- Nucleoproteins: Combination of protein and nucleic acid
- Mucoproteins: Combination of proteins and carbohydrates (>4%)
- Glycoproteins: Combination of proteins and carbohydrates(<4%)
- Chromoproteins: Combination of proteins and coloured pigments.
- Lipoproteins: Combination of proteins and lipids.
- Metalloprotein: Combination of proteins and metal ions.
- Phosphoprotein: Combination of proteins and phosphate group.
- Derived proteins: When proteins are hydrolyzed by acids, alkalies or enzymes, the degradation products obtained from them are called derived proteins.
On the basis of nature of Molecules
- Acidic proteins: They exist as anion and contain acidic amino acids. e.g. blood groups.
- Basic proteins: They exist as cations and are rich in basic amino acids e.g. lysine, arginine etc.
Functions of Proteins
- Structural functions: Proteins are called as the building blocks of the body. They are an essential component of various structures in the cell and tissues. We also find these proteins in the outer membrane of all cells in the human body. We can also find structural proteins in hair, skin, and muscles. Proteins often act to strengthen these structures. Proteins working together can allow movement within the body, such as contraction of muscles and movement of food through the digestive system etc. They are needed for the growth, development, healing, and repair of tissues.
- Protective: Proteins are the main constituent of antibodies that protect our body against antigens and pathogens thus preventing infections.
- Hormonal regulation: Hormones are majorly composed of proteins. Hormones play a vital role in regulating muscle mass, sex hormones, and growth and development.
- Enzymes: Proteins are called as biological buffers because they, as enzymes, regulate many different biochemical reactions that are occurring in the body.
Solved Example for You
Q: Peptide bond form between two amino acids through
(a) Addition of water (b) Loss of water
(c) Decarboxylation (d) Deamination
Sol. (b) loss of water.
The formation of a peptide bond is a dehydration reaction where a molecule of water is released. Therefore, the correct answer is the option (b).
Customize your course in 30 seconds
Which class are you in.

Biomolecules
- Polysaccharides
- Bond Linking Monomers
- Nucleic Acids
- Metabolic Basis For Living
- Biomacromolecules
One response to “Biomacromolecules”
DNA consists of thymine or thiamine??
Leave a Reply Cancel reply
Your email address will not be published. Required fields are marked *
Download the App


- school Campus Bookshelves
- menu_book Bookshelves
- perm_media Learning Objects
- login Login
- how_to_reg Request Instructor Account
- hub Instructor Commons
- Download Page (PDF)
- Download Full Book (PDF)
- Periodic Table
- Physics Constants
- Scientific Calculator
- Reference & Cite
- Tools expand_more
- Readability
selected template will load here
This action is not available.

6.4: Protein Synthesis
- Last updated
- Save as PDF
- Page ID 27396

- Suzanne Wakim & Mandeep Grewal
- Butte College
The Central Dogma of Biology
Your DNA , or deoxyribonucleic acid, contains the genes that determine who you are. How can this organic molecule control your characteristics? DNA contains instructions for all the proteins your body makes. Proteins , in turn, determine the structure and function of all your cells. What determines a protein ’s structure? It begins with the sequence of amino acids that make up the protein. Instructions for making proteins with the correct sequence of amino acids are encoded in DNA.

DNA is found in chromosomes. In eukaryotic cells, chromosomes always remain in the nucleus, but proteins are made at ribosomes in the cytoplasm or on the rough endoplasmic reticulum (RER) . How do the instructions in DNA get to the site of protein synthesis outside the nucleus? Another type of nucleic acid is responsible. This nucleic acid is RNA or ribonucleic acid. RNA is a small molecule that can squeeze through pores in the nuclear membrane. It carries the information from DNA in the nucleus to a ribosome in the cytoplasm and then helps assemble the protein. In short:
DNA → RNA → Protein
Discovering this sequence of events was a major milestone in molecular biology. It is called the central dogma of biology . The two processes involved in the central dogma are transcription and translation.
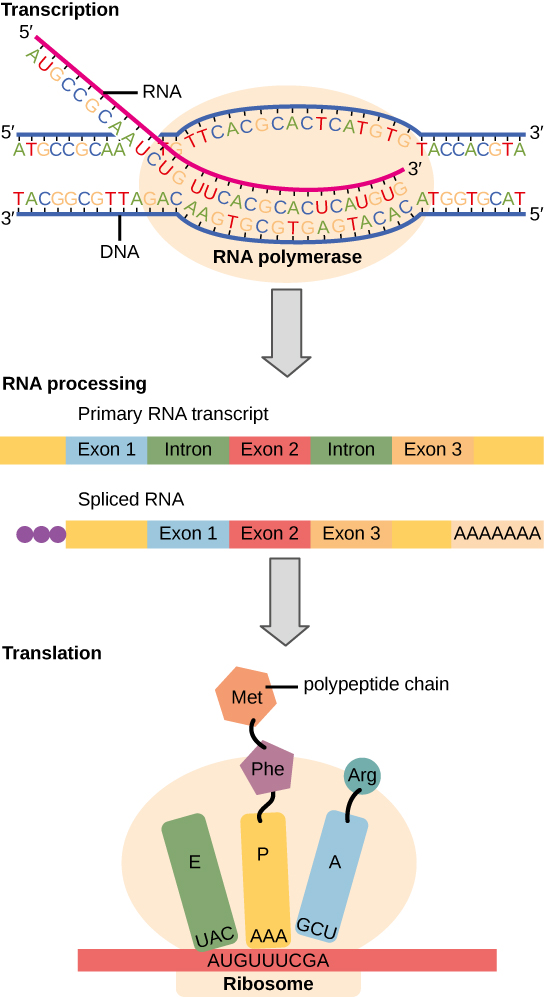
Transcription
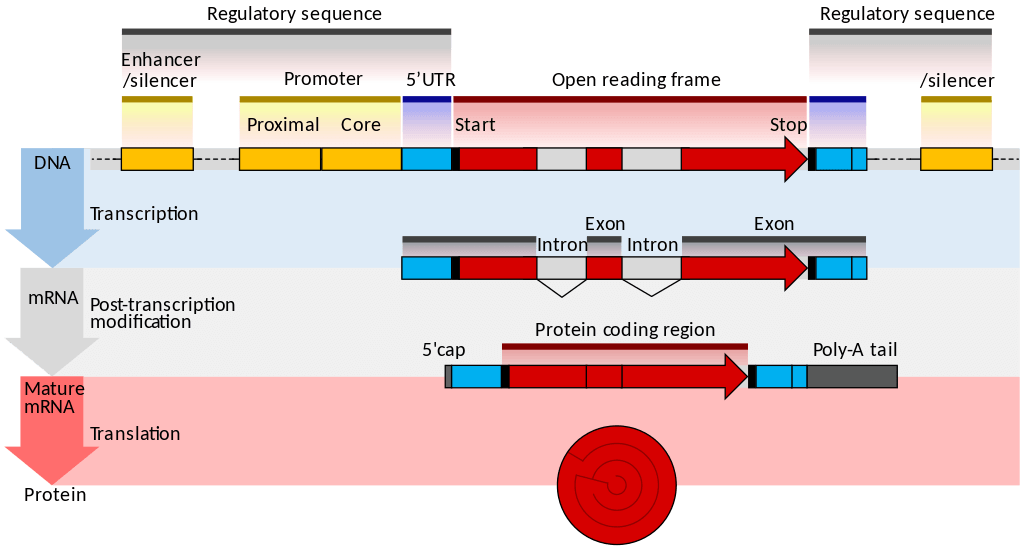
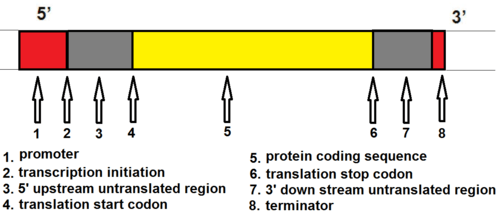
Transcription is the first part of the central dogma of molecular biology: DNA → RNA . It is the transfer of genetic instructions in DNA to mRNA. Transcription happens in the nucleus of the cell. During transcription, a strand of mRNA is made that is complementary to a strand of DNA called a gene. A gene can easily be identified from the DNA sequence. A gene contains the basic three regions, promoter, coding sequence (reading frame), and terminator. There are more parts of a gene which are illustrated in Figure \(\PageIndex{3}\).
Steps of Transcription
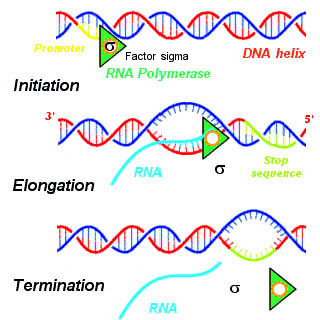
Transcription takes place in three steps, called initiation, elongation, and termination. The steps are illustrated in Figure \(\PageIndex{4}\).
- Initiation is the beginning of transcription. It occurs when the enzyme RNA polymerase binds to a region of a gene called the promoter . This signals the DNA to unwind so the enzyme can “read” the bases in one of the DNA strands. The enzyme is ready to make a strand of mRNA with a complementary sequence of bases. The promoter is not part of the resulting mRNA
- Elongation is the addition of nucleotides to the mRNA strand.
Processing mRNA
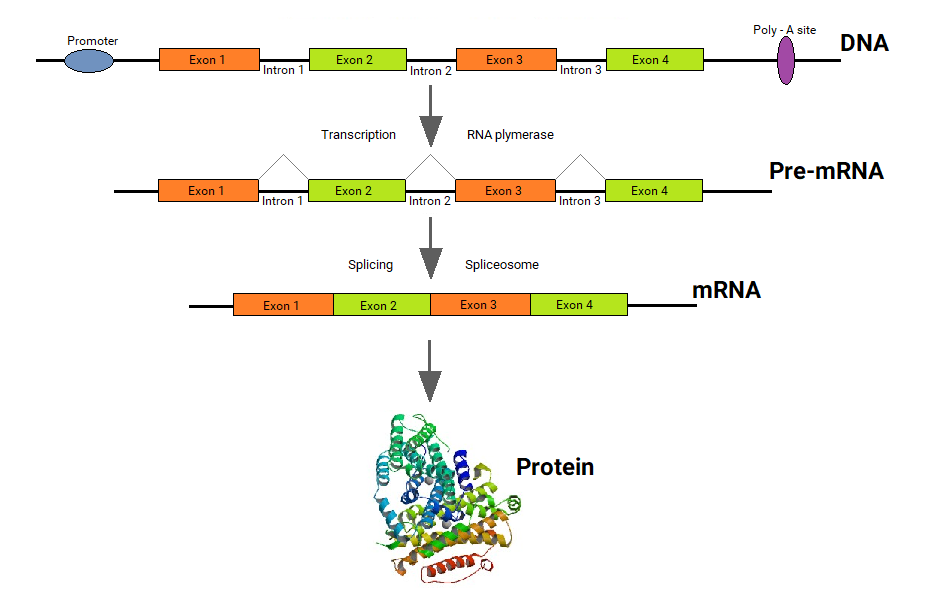
In eukaryotes, the new mRNA is not yet ready for translation. At this stage, it is called pre-mRNA, and it must go through more processing before it leaves the nucleus as mature mRNA. The processing may include the addition of a 5' cap, splicing, editing, and 3' polyadenylation (poly-A) tail. These processes modify the mRNA in various ways. Such modifications allow a single gene to be used to make more than one protein. See Figure \(\PageIndex{5}\) as you read below:
- 5' cap protects mRNA in the cytoplasm and helps in the attachment of mRNA with the ribosome for translation.
- Splicing removes introns from the protein-coding sequence of mRNA. Introns are regions that do not code for the protein. The remaining mRNA consists only of regions called exons that do code for the protein.
- Editing changes some of the nucleotides in mRNA. For example, a human protein called APOB, which helps transport lipids in the blood, has two different forms because of editing. One form is smaller than the other because editing adds an earlier stop signal in mRNA.
- Polyadenylation adds a “tail” to the mRNA. The tail consists of a string of As (adenine bases). It signals the end of mRNA. It is also involved in exporting mRNA from the nucleus, and it protects mRNA from enzymes that might break it down.
Translation
The translation is the second part of the central dogma of molecular biology: RNA --> Protein . It is the process in which the genetic code in mRNA is read to make a protein. The translation is illustrated in Figure \(\PageIndex{6}\). After mRNA leaves the nucleus, it moves to a ribosome, which consists of rRNA and proteins. Translation happens on the ribosomes floating in the cytosol, or on the ribosomes attached to the rough endoplasmic reticulum. The ribosome reads the sequence of codons in mRNA, and molecules of tRNA bring amino acids to the ribosome in the correct sequence.
To understand the role of tRNA, you need to know more about its structure. Each tRNA molecule has an anticodon for the amino acid it carries. An anticodon is complementary to the codon for an amino acid. For example, the amino acid lysine has the codon AAG, so the anticodon is UUC. Therefore, lysine would be carried by a tRNA molecule with the anticodon UUC. Wherever the codon AAG appears in mRNA, a UUC anticodon of tRNA temporarily binds. While bound to mRNA, tRNA gives up its amino acid. With the help of rRNA, bonds form between the amino acids as they are brought one by one to the ribosome, creating a polypeptide chain. The chain of amino acids keeps growing until a stop codon is reached.
Ribosomes, which are just made out of rRNA (ribosomal RNA) and protein, have been classified as ribozymes because the rRNA has enzymatic activity. The rRNA is important for the peptidyl transferase activity that bonds amino acids. Ribosomes have two subunits of rRNA and protein. The large subunit has three active sites called E, P, and A sites. These sites are important in the catalytic activity of ribosomes.
Just as with mRNA synthesis, protein synthesis can be divided into three phases: initiation, elongation, and termination. In addition to the mRNA template, many other molecules contribute to the process of translation, such as ribosomes, tRNAs, and various enzymatic factors
Translation Initiation: The small subunit binds to a site upstream (on the 5' side) of the start of the mRNA. It proceeds to scan the mRNA in the 5'-->3' direction until it encounters the START codon (AUG). The large subunit attaches and the initiator tRNA, which carries methionine (Met), binds to the P site on the ribosome.
Translation Elongation: The ribosome shifts one codon at a time, catalyzing each process that occurs in the three sites. With each step, a charged tRNA enters the complex, the polypeptide becomes one amino acid longer, and an uncharged tRNA departs. The energy for each bond between amino acids is derived from GTP, a molecule similar to ATP. Briefly, the ribosomes interact with other RNA molecules to make chains of amino acids called polypeptide chains, due to the peptide bond that forms between individual amino acids. Inside the ribosome, three sites participate in the translation process, the A, P, and E sites. Amazingly, the E. coli translation apparatus takes only 0.05 seconds to add each amino acid, meaning that a 200-amino acid polypeptide could be translated in just 10 seconds.
Translation Termination : Termination of translation occurs when a stop codon (UAA, UAG, or UGA) is encountered (see Figure \(\PageIndex{7}\). When the ribosome encounters the stop codon, the growing polypeptide is released with the help of various releasing factors and the ribosome subunits dissociate and leave the mRNA. After many ribosomes have completed translation, the mRNA is degraded so the nucleotides can be reused in another transcription reaction.
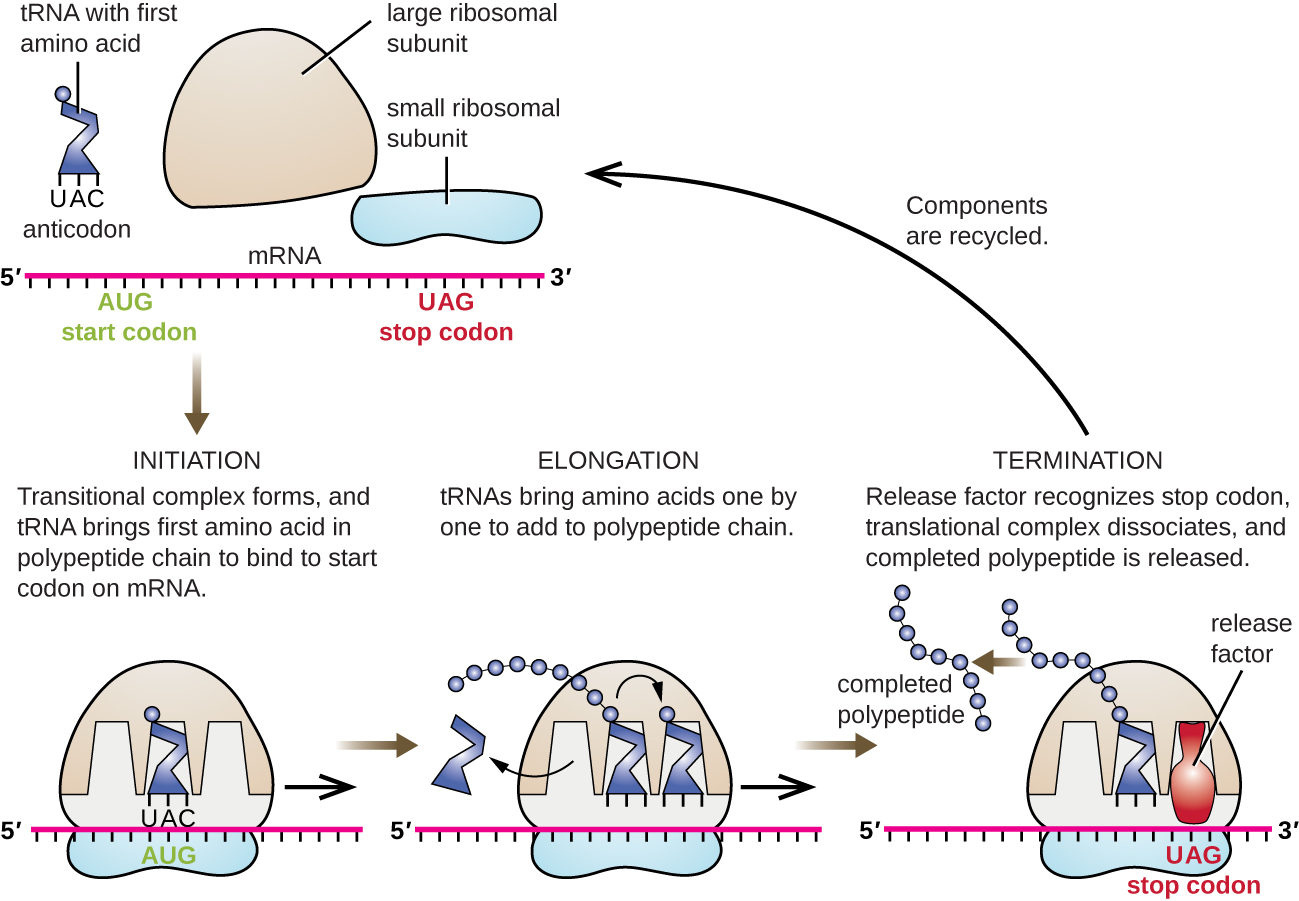
What Happens Next?
After a polypeptide chain is synthesized, it may undergo additional processes. For example, it may assume a folded tertiary shape due to interactions among its amino acids. It may also bind with other polypeptides or with different types of molecules, such as lipids or carbohydrates. Many proteins travel to the Golgi apparatus within the cytoplasm to be modified for the specific job they will do.
Summary of Central Dogma
- Relate protein synthesis and its two major phases to the central dogma of molecular biology.
- Identify the steps of transcription, and summarize what happens during each step.
- Explain how mRNA is processed before it leaves the nucleus.
- Describe what happens during the translation phase of protein synthesis.
- What additional processes may a polypeptide chain undergo after it is synthesized?
- Where does transcription take place in eukaryotes?
- Where does translation take place?
- Contains the codons
- Contains the anticodons
- Makes up the ribosome, along with proteins
- What is the complementary sequence on the other DNA strand?
- What is the complementary sequence in the mRNA? What is this sequence called?
- @hat is the resulting sequence in the tRNA? What is this sequence called? What do you notice about this sequence compared to the original DNA triplet on the template strand?
- Both A and B
- True or False. Introns in mRNA bind to tRNA at the ribosome.
- True or False. tRNAs can be thought of as the link between amino acids and codons in the mRNA.
Explore More
Messenger RNA molecules are "spliced" in order to create the mRNA involved in protein synthesis. Learn the process here:
Attributions
- How proteins are made by Nicolle Rager, National Science Foundation, public domain via Wikimedia Commons
- Gene structure eukaryote by Thomas Shafee , licensed CC BY 4.0 via Wikimedia Commons
- Components of a gene by Mandeep Grewal, CC BY 4.0
- Transcription by Calibuon , released into the public domain via Wikimedia Commons
- Transcript and splicing by Ganeshmanohar , CC BY-SA 4.0 via Wikimedia Commons
- Initiation and elongation by Jordan Nguyen, CC BY-SA 4.0 via Wikimedia Commons
- Protein synthesis by OpenStax, CC BY 4.0
- Gene regulation by OpenStax, CC BY 4.0
- Text adapted from Human Biology by CK-12 licensed CC BY-NC 3.0
Assigning secondary structure in proteins using AI
- Original Paper
- Published: 17 August 2021
- Volume 27 , article number 252 , ( 2021 )
Cite this article
- Jisna Vellara Antony ORCID: orcid.org/0000-0001-5210-9583 1 ,
- Prayagh Madhu 2 ,
- Jayaraj Pottekkattuvalappil Balakrishnan ORCID: orcid.org/0000-0002-9924-9046 1 &
- Hemant Yadav 1
466 Accesses
4 Citations
Explore all metrics
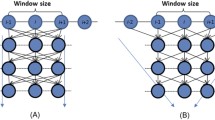
Knowledge about protein structure assignment enriches the structural and functional understanding of proteins. Accurate and reliable structure assignment data is crucial for secondary structure prediction systems. Since the 1980s, various methods based on hydrogen bond analysis and atomic coordinate geometry, followed by machine learning, have been employed in protein structure assignment. However, the assignment process becomes challenging when missing atoms are present in the protein files. Our method proposed a multi-class classifier program named DLFSA for assigning protein secondary structure elements (SSE) using convolutional neural networks (CNNs). A fast and efficient GPU-based parallel procedure extracts fragments from protein files. The model implemented in this work is trained with a subset of the protein fragments and achieves 88.1% and 82.5% train and test accuracy, respectively. The model uses only C α coordinates for secondary structure assignments. The model has been successfully tested on a few full-length proteins also. Results from the fragment-based studies demonstrate the feasibility of applying deep learning solutions for structure assignment problems.
This is a preview of subscription content, log in via an institution to check access.
Access this article
Price includes VAT (Russian Federation)
Instant access to the full article PDF.
Rent this article via DeepDyve
Institutional subscriptions
Similar content being viewed by others
Protein secondary structure assignment using residual networks
Jisna Vellara Antony, Roosafeed Koya, … Jayaraj Pottekkattuvalappil Balakrishnan
Protein Secondary Structure Prediction Using Deep Convolutional Neural Fields
Sheng Wang, Jian Peng, … Jinbo Xu
Energy Profile Bayes and Thompson Optimized Convolutional Neural Network protein structure prediction
Varanavasi Nallasamy & Malarvizhi Seshiah
Data availability
DLFSA is made available to the public through the Web portal— www.proteinallinfo.in . The datasets generated during and/or analyzed during the current study are not publicly available due to their large size but are available from the corresponding author on reasonable request.
Pauling L, Corey RB, Branson HR (1951) The structure of proteins: two hydrogen-bonded helical configurations of the polypeptide chain. Proc Natl Acad Sci 37(4):205–211
Article CAS PubMed PubMed Central Google Scholar
Reeb J, Rost B (2019) Secondary structure prediction. Encyclopedia of Bioin-formatics and Computational Biology, pp 488–496
Chapter Google Scholar
Srinivasan R, Rose GD (1999) A physical basis for protein secondary structure. Proc Natl Acad Sci 96(25):14258–14263
Eisenberg D (2003) The discovery of the α-helix and β-sheet, the principal structural features of proteins. Proc Natl Acad Sci 100(20):11207–11210
Zhou J, Wang H, Zhao Z, Xu R, Lu Q (2018) CNNH_PSS: protein 8-class secondary structure prediction by convolutional neural network with highway. BMC Bioinform 19(4):99–109
Google Scholar
Abbass J, Nebel JC, Mansour N, Elloumi M, Zomaya AY (2013) Ab initio protein structure prediction: methods and challenges. Biol Knowl Discov Handb. John Wiley & Sons, Inc, Hoboken, New Jersey, pp 703–724
Anfinsen CB (1973) Principles that govern the folding of protein chains. Science 181(4096):223–230
Article CAS PubMed Google Scholar
Onuchic JN, Wolynes PG (2004) Theory of protein folding. Curr Opin Struct Biol 14(1):70–75
Kabsch W, Sander C (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22(12):2577–2637
Frishman D, Argos P (1995) Knowledge-based protein secondary structure assignment. Proteins Struct Funct Bioinf 23(4):566–579
Article CAS Google Scholar
Ramachandran GT, Sasisekharan V (1968) Conformation of polypeptides and proteins. Adv Protein Chem 23:283–437
Zacharias J, Knapp EW (2014) Protein secondary structure classification revisited: processing DSSP information with PSSC. J Chem Inf Model 54(7):2166–2179
Fodje MN, Al-Karadaghi S (2002) Occurrence, conformational features and amino acid propensities for the π-helix. Protein Eng Des Sel 15(5):353–358
Nagy G, Oostenbrink C (2014) Dihedral-based segment identification and classification of biopolymers I: proteins. J Chem Inf Model 54(1):266–277
Cubellis MV, Cailliez F, Lovell SC (2005) Secondary structure assignment that accurately reflects physical and evolutionary characteristics. BMC Bioinform 6(4):1–9
Richards FM, Kundrot CE (1988) Identification of structural motifs from protein coordinate data: secondary structure and first-level supersecondary structure. Proteins Struct Funct Bioinf 3(2):71–84
Sklenar H, Etchebest C, Lavery R (1989) Describing protein structure: a general algorithm yielding complete helicoidal parameters and a unique overall axis. Proteins Struct Funct Bioinf 6(1):46–60
Hosseini SR, Sadeghi M, Pezeshk H, Eslahchi C, Habibi M (2008) PROSIGN: a method for protein secondary structure assignment based on three-dimensional coordinates of consecutive Cα atoms. Comput Biol Chem 32(6):406–411
Labesse G, Colloc'h N, Pothier J, Mornon JP (1997) P-SEA: a new efficient assignment of secondary structure from Cα trace of proteins. Bioinformatics 13(3):291–295
Majumdar I, Krishna SS, Grishin NV (2005) PALSSE: a program to delineate linear secondary structural elements from protein structures. BMC Bioinform 6(1):1–24
Taylor WR (2001) Defining linear segments in protein structure. J Mol Biol 310(5):1135–1150
Dupuis F, Sadoc JF, Mornon JP (2004) Protein secondary structure assignment through Voronoi tessellation. Proteins Struct Funct Bioinf 55(3):519–528
Park SY, Yoo MJ, Shin JM, Cho KH (2011) SABA (secondary structure assignment program based on only alpha carbons): a novel pseudo center geometrical criterion for accurate assignment of protein secondary structures. BMB Rep 44(2):118–122
Zhang W, Dunker AK, Zhou Y (2008) Assessing secondary structure assignment of protein structures by using pairwise sequence-alignment benchmarks. Proteins Struct Funct Bioinf 71(1):61–67
Cao C, Wang G, Liu A, Xu S, Wang L, Zou S (2016) A new secondary structure assignment algorithm using Cα backbone fragments. Int J Mol Sci 17(3):333
Article PubMed PubMed Central CAS Google Scholar
Konagurthu AS, Lesk AM, Allison L (2012) Minimum message length inference of secondary structure from protein coordinate data. Bioinformatics 28(12):i97–i105
Haghighi H, Higham J, Henchman RH (2016) Parameter-free hydrogen-bond definition to classify protein secondary structure. J Phys Chem B 120(33):8566–8570
Kumar P, Bansal M (2012) HELANAL-Plus: a web server for analysis of helix geometry in protein structures. J Biomol Struct Dyn 30(6):773–783
King SM, Johnson WC (1999) Assigning secondary structure from protein coordinate data. Proteins Struct Funct Bioinf 35(3):313–320
Carter P, Andersen CA, Rost B (2003) DSSPcont: continuous secondary structure assignments for proteins. Nucleic Acids Res 31(13):3293–3295
Konagurthu AS, Allison L, Stuckey PJ, Lesk AM (2011) Piecewise linear approximation of protein structures using the principle of minimum message length. Bioinformatics 27(13):i43–i51
Levitt M, Greer J (1977) Automatic identification of secondary structure in globular proteins. J Mol Biol 114(2):181–239
Cao C, Xu S, Wang L (2015) An algorithm for protein helix assignment using helix geometry. PLoS One 10(7):e0129674
Klose DP, Wallace BA, Janes RW (2010) 2Struc: the secondary structure server. Bioinformatics 26(20):2624–2625
Kumar P, Bansal M (2015) Identification of local variations within secondary structures of proteins. Acta Crystallogr D Biol Crystallogr 71(5):1077–1086
Habibia M, Eslahchia C, Pezeshkc H, Sadeghid M (2008) An information-theoretic approach to secondary structure assignment, Journal of Science (University of Tehran) (JSUT)
Taylor T, Rivera M, Wilson G, Vaisman II (2005) New method for protein secondary structure assignment based on a simple topological descriptor. Proteins Struct Funct Bioinf 60(3):513–524
Zhang Y, Sagui C (2015) Secondary structure assignment for conformationally irregular peptides: comparison between DSSP, STRIDE and KAKSI. J Mol Graph Model 55:72–84
Article PubMed CAS Google Scholar
Law SM, Frank AT, Brooks III CL (2014) PCASSO: a fast and efficient Cα-based method for accurately assigning protein secondary structure elements. J Comput Chem 35(24):1757–1761
Salawu EO (2016) RaFoSA: Random forests secondary structure assignment for coarse-grained and all-atom protein systems. Cogent Biol 2(1):1214061
Wang J, Cao H, Zhang JZ, Qi Y (2018) Computational protein design with deep learning neural networks. Sci Rep 8(1):1–9
Cheng J, Tegge AN, Baldi P (2008) Machine learning methods for protein structure prediction. IEEE Rev Biomed Eng 1:41–49
Article PubMed Google Scholar
Zhang B, Li J, Lü Q (2018) Prediction of 8-state protein secondary structures by a novel deep learning architecture. BMC Bioinform 19(1):1–13
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521(7553):436–444
Goh GB, Hodas NO, Vishnu A (2017) Deep learning for computational chemistry. J Comput Chem 38(16):1291–1307
O'Shea, K., & Nash, R. (2015). An introduction to convolutional neural networks. arXiv preprint arXiv:1511.08458.
Busia, A., Collins, J., & Jaitly, N. (2016). Protein secondary structure prediction using deep multi-scale convolutional neural networks and next-step conditioning. arXiv preprint arXiv:1611.01503.
Zamora-Resendiz R, Crivelli S (2019) Structural learning of proteins using graph convolutional neural networks. bioRxiv, 610444, Cold Spring Harbor Laboratory
Niepert, M., Ahmed, M., & Kutzkov, K. (2016). Learning convolutional neural networks for graphs. In International conference on machine learning (pp. 2014-2023). PMLR.
https://www.rcsb.org/structure/ , accessed : 2020-09-09.
Holmes JB, Tsai J (2004) Some fundamental aspects of building protein structures from fragment libraries. Protein Sci 13(6):1636–1650
Xu D, Zhang Y (2013) Toward optimal fragment generations for ab initio protein structure assembly. Proteins Struct Funct Bioinf 81(2):229–239
de Oliveira SH, Shi J, Deane CM (2015) Building a better fragment library for de novo protein structure prediction. PLoS One 10(4):e0123998
Abbass J, Nebel JC (2015) Customised fragments libraries for protein structure prediction based on structural class annotations. BMC Bioinform 16(1):1–13
Trevizani R, Custódio FL, Dos Santos KB, Dardenne LE (2017) Critical features of fragment libraries for protein structure prediction. PLoS One 12(1):e0170131
Abbass J, Nebel JC (2020) Enhancing fragment-based protein structure prediction by customising fragment cardinality according to local secondary structure. BMC Bioinform 21:1–23
https://www.djangoproject.com/ , accessed : 2020-12-12.
Download references
Acknowledgements
The authors would like to thank the Central Computing Centre, National Institute of Technology Calicut (NITC), for providing GPU servers for this work. The authors would like to acknowledge the valuable suggestions from Jinto Antony for developing the Web portal.
Code availability
The library generation source codes and the model codes are made open at https://github.com/jisnava/DLFSA/ .
It is part of my (VA Jisna) PhD work at the National Institute of Technology Calicut, India. The research is funded by the Ministry of Human Resource Development, India.
Author information
Authors and affiliations.
Department of Computer Science and Engineering, National Institute of Technology Calicut, Kerala, 673601, India
Jisna Vellara Antony, Jayaraj Pottekkattuvalappil Balakrishnan & Hemant Yadav
Computer Science and Engineering Dept., Rajiv Gandhi Institute of Technology, Kottayam, India
Prayagh Madhu
You can also search for this author in PubMed Google Scholar
Contributions
Jisna Vellara Antony (JVA) did the conceptualization. Hemant Yadav developed the fragment library. Prayagh Madhu did the model coding. JVA developed the Web portal and wrote the manuscript. Jayaraj Pottekkattuvalappil Balakrishnan supervised the project. All authors read and approved the final manuscript.
Corresponding author
Correspondence to Jisna Vellara Antony .
Ethics declarations
Competing interests.
The authors declare no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
(DOCX 335 kb)
Rights and permissions
Reprints and permissions
About this article
Antony, J.V., Madhu, P., Balakrishnan, J.P. et al. Assigning secondary structure in proteins using AI. J Mol Model 27 , 252 (2021). https://doi.org/10.1007/s00894-021-04825-x
Download citation
Received : 22 January 2021
Accepted : 16 June 2021
Published : 17 August 2021
DOI : https://doi.org/10.1007/s00894-021-04825-x
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Protein structure assignment
- Deep learning
- Fragment library creation
- Convolutional neural networks
- Protein fragments
- Protein secondary structures
- Multi-class classifier
- Find a journal
- Publish with us
- Track your research
- Protein Structure And Levels Of Protein
Protein Structure
Protein structures are made by condensation of amino acids forming peptide bonds. The sequence of amino acids in a protein is called its primary structure. The secondary structure is determined by the dihedral angles of the peptide bonds, the tertiary structure by the folding of protein chains in space. Association of folded polypeptide molecules to complex functional proteins results in quaternary structure.

Define Protein Structure
Protein structure is defined as a polymer of amino acids joined by peptide bonds.

Related Topics
- Amino acids
- Peptide bond
- Denaturation of Proteins
Let us see how a peptide bond is established from the following reaction:
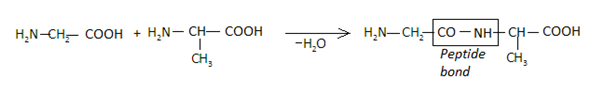
Formation of Peptide Bond
We can thus see that the peptide bond (-CO-NH) is formed between the amine group of one molecule and the carboxyl group of the adjacent molecule followed by the elimination of a water molecule. This bond is otherwise an amide linkage. When peptide bonds are established among more than ten amino acids, they together form a polypeptide chain. Very often, when a polypeptide chain has a mass exceeding 10000u and the number of amino acids in the chain exceeding 100, we get a protein.
Also Read: Denaturation of Proteins
Recommended Videos

Classification of Proteins
Based on the molecular shape, proteins can be classified into two types.
1. Fibrous Proteins:
When the polypeptide chains run parallel and are held together by hydrogen and disulfide bonds, then the fiber-like structure is formed. Such proteins are generally insoluble in water. These are water-insoluble proteins.
Example – keratin (present in hair, wool, and silk) and myosin (present in muscles), etc.
2. Globular Proteins:
This structure results when the chains of polypeptides coil around to give a spherical shape. These are usually soluble in water.
Example – Insulin and albumins are common examples of globular proteins.
Levels of Protein Structure
1. primary structure of protein.
- The Primary structure of proteins is the exact ordering of amino acids forming their chains.
- The exact sequence of the proteins is very important as it determines the final fold and therefore the function of the protein.
- The number of polypeptide chains together form proteins. These chains have amino acids arranged in a particular sequence which is characteristic of the specific protein. Any change in the sequence changes the entire protein.
The following picture represents the primary protein structure (an amino acid chain). As you might expect, the amino acid sequence within the polypeptide chain is crucial for the protein’s proper functioning. This sequence is encrypted in the DNA genetic code. If mutation is present in the DNA and the amino acid sequence is changed, the protein function may be affected.

The protein ‘s primary structure is the amino acid sequence in its polypeptide chain. If proteins were popcorn stringers designed to decorate a Christmas tree, a protein ‘s primary structure is the sequence in which various shapes and varieties of popped maize are strung together.
Covalent, peptide bonds which connect the amino acids together maintain the primary structure of a protein.
All documented genetic disorders, such as cystic fibrosis, sickle cell anemia, albinism, etc., are caused by mutations resulting in alterations in the primary protein structures, which in turn lead to alterations in the secondary , tertiary and probably quarterly structure.
Amino acids are small organic molecules consisting of a chiral carbon with four substituents. Of those only the fourth the side chain is different among amino acids.
2. Secondary Structure of Protein
Secondary structure of protein refers to local folded structures that form within a polypeptide due to interactions between atoms of the backbone.
- The proteins do not exist in just simple chains of polypeptides.
- These polypeptide chains usually fold due to the interaction between the amine and carboxyl group of the peptide link.
- The structure refers to the shape in which a long polypeptide chain can exist.
- They are found to exist in two different types of structures α – helix and β – pleated sheet structures.
- This structure arises due to the regular folding of the backbone of the polypeptide chain due to hydrogen bonding between -CO group and -NH groups of the peptide bond.
- However, segments of the protein chain may acquire their own local fold, which is much simpler and usually takes the shape of a spiral an extended shape or a loop. These local folds are termed secondary elements and form the proteins secondary structure.

(a) α – Helix:
α – Helix is one of the most common ways in which a polypeptide chain forms all possible hydrogen bonds by twisting into a right-handed screw with the -NH group of each amino acid residue hydrogen-bonded to the -CO of the adjacent turn of the helix. The polypeptide chains twisted into a right-handed screw.
(b) β – pleated sheet:
In this arrangement, the polypeptide chains are stretched out beside one another and then bonded by intermolecular H-bonds. In this structure, all peptide chains are stretched out to nearly maximum extension and then laid side by side which is held together by intermolecular hydrogen bonds. The structure resembles the pleated folds of drapery and therefore is known as β – pleated sheet
3. Tertiary Structure of Protein
- This structure arises from further folding of the secondary structure of the protein.
- H-bonds, electrostatic forces, disulphide linkages, and Vander Waals forces stabilize this structure.
- The tertiary structure of proteins represents overall folding of the polypeptide chains, further folding of the secondary structure.
- It gives rise to two major molecular shapes called fibrous and globular.
- The main forces which stabilize the secondary and tertiary structures of proteins are hydrogen bonds, disulphide linkages, van der Waals and electrostatic forces of attraction.

4. Quaternary Structure of Protein
The spatial arrangement of various tertiary structures gives rise to the quaternary structure. Some of the proteins are composed of two or more polypeptide chains referred to as sub-units. The spatial arrangement of these subunits with respect to each other is known as quaternary structure.

The exact amino acid sequence of each protein drives it to fold into its own unique and biologically active three-dimensional fold also known as the tertiary structure. Proteins consist of different combinations of secondary elements some of which are simple whereas others are more complex. Parts of the protein chain, which have their own three-dimensional fold and can be attributed to some function are called “domains” . These are considered today as the evolutionary and functional building blocks of proteins.
Many proteins, most of which are enzymes contain organic or elemental components needed for their activity and stability. Thus the study of protein evolution not only gives structural insight but also connects proteins of quite different parts of the metabolism.
Also Read: Laboratory Test of Proteins
Rules of Protein Structure
- The type determines the function of a protein.
- A protein’s shape is determined by its primary structure (the amino acid sequence).
- The amino acid sequence within a protein is determined by the encoding sequence of nucleotides in the gene (DNA).
Summary of Protein Structure
Linderstrom-Lang (1952) in particular first suggested a hierarchy of protein structure with four levels: central, secondary, tertiary , and quaternary. You are already familiar with this hierarchy, because the most useful starting point for teaching basic protein structure is this structural grouping.
- The primary structure of protein is the hierarchy’s basic level, and is the particular linear sequence of amino acids comprising one polypeptide chain.
- Secondary structure is the next level up from the primary structure, and is the regular folding of regions into specific structural patterns within one polypeptide chain. Hydrogen bonds between the carbonyl oxygen and the peptide bond amide hydrogen are normally held together by secondary structures.
- Tertiary structure is the next level up from the secondary structure, and is the particular three-dimensional arrangement of all the amino acids in a single polypeptide chain. This structure is usually conformational, native, and active, and is held together by multiple noncovalent interactions.
- Quaternary structure is the next ‘step up’ between two or more polypeptide chains from the tertiary structure and is the specific spatial arrangement and interactions.
Frequently Asked Questions – FAQs
What makes up protein structure.
A protein’s primary structure refers to the amino acid sequence in the polypeptide chain. Peptide bonds that are made during the protein biosynthesis process hold the primary structure together.
What are the 4 stages of protein structure?
Four levels of structure of proteins. The principal, secondary, tertiary and quaternary levels of protein structure are the four stages. To fully understand how a protein functions, it is helpful to understand the purpose and role of each level of protein structure.
What is the process of protein folding?
The folding of proteins is the mechanism through which a protein structure assumes its functional shape or conformation. Both molecules of protein are heterogeneous unbranched amino acid chains. They may perform their biological function by coiling and folding in a particular three-dimensional shape.
How proteins are formed?
Amino acids form a polypeptide, In another words when amino acids bound by a sequence of peptide bonds , leads to formation of proteins. The polypeptide then folds into a particular conformation based on the interactions (strained lines) between its side chains of amino acids.
Is DNA a protein?
DNA is often associated with proteins in the nucleus called histones, but DNA itself is not a protein. No. DNA is a nucleic acid consisting of phosphate and sugar groups, bases ( purines and pyrimidines), while proteins are large molecules made up of one or more long amino acid chains.
What stabilizes protein structure?
Hydrogen bonding in the polypeptide chain and between amino acid “R” groups helps to preserve protein structure by keeping the protein in the form formed by the hydrophobic interactions. What is called a disulfide bridge is formed by this sort of bonding.
What determines protein structure?
In the polypeptide chain, the main structure of a protein relates to the amino acid sequence. The primary structure is bound together by peptide bonds that are made during the phase of protein biosynthesis. The primary structure of a protein is determined by the gene corresponding to the protein.
What is the primary structure of a protein?
The linear sequence of amino acids within a protein is called the primary structure of the protein. A sequence of just twenty amino acids, each of which has a special side chain, is made up of proteins. The side chains of amino acids are chemically distinct.

Put your understanding of this concept to test by answering a few MCQs. Click ‘Start Quiz’ to begin!
Select the correct answer and click on the “Finish” button Check your score and answers at the end of the quiz
Visit BYJU’S for all Chemistry related queries and study materials
Your result is as below
Request OTP on Voice Call
Leave a Comment Cancel reply
Your Mobile number and Email id will not be published. Required fields are marked *
Post My Comment
Helped a lot Thank you Byjus
- Share Share
Register with BYJU'S & Download Free PDFs
Register with byju's & watch live videos.

Browse Course Material
Course info, instructors.
- Prof. Christopher Burge
- Prof. David Gifford
- Prof. Ernest Fraenkel
Departments
- Biological Engineering
- Electrical Engineering and Computer Science
- Health Sciences and Technology
As Taught In
- Computational Biology
- Computational Modeling and Simulation
- Computation and Systems Biology
Learning Resource Types
Foundations of computational and systems biology, lecture 13: predicting protein structure.
Description: This lecture on predicting protein structure covers refining a partially correct structure. Methods include energy minimization, molecular dynamics, and simulated annealing. He moves on to methods for predicting structure from a sequence of amino acid.
Instructor: Prof. Ernest Fraenkel
- Download video
- Download transcript

You are leaving MIT OpenCourseWare
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
Proteins articles from across Nature Portfolio
Proteins are biopolymers of amino acids (polypeptides), joined by peptide bonds, that are generated by ribosomes. The amino acid sequence, encoded by its gene, determines a protein’s structure and function. Newly synthesized proteins can be modified by post-translational modification, altering its protein folding, stability and activity. Proteins often associate into protein–protein interaction networks for function.
Related Subjects
- Acetyltransferases
- Blood proteins
- Carrier proteins
- Cell-cycle proteins
- Cerebrospinal fluid proteins
- Circadian rhythm signalling peptides and proteins
- Contractile proteins
- Cytoskeletal proteins
- DNA-binding proteins
- G protein-coupled receptors
- Glycoproteins
- GTP-binding protein regulators
- Intracellular signalling peptides and proteins
- Lipoproteins
- Membrane proteins
- Metalloproteins
- Mitochondrial proteins
- Nuclear receptors
- Nucleoproteins
- Nucleotide-binding proteins
- Oncogene proteins
- Phosphoproteins
- Phosphorylases
- Ribosomal proteins
- RNA-binding proteins
- Scleroproteins
- Selenium-binding proteins
- Sumoylated proteins
- Thioredoxins
- Transcription factors
- Tumour-suppressor proteins
- Ubiquitylated proteins
- Viral proteins
Latest Research and Reviews
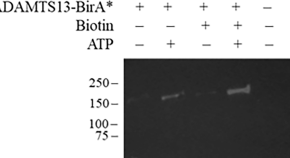
Optimization of plasma-based BioID identifies plasminogen as a ligand of ADAMTS13
- Hasam Madarati
- Veronica DeYoung
- Colin A. Kretz
Type 1 secretion necessitates a tight interplay between all domains of the ABC transporter
- Manuel T. Anlauf
- Florestan L. Bilsing
- Lutz Schmitt
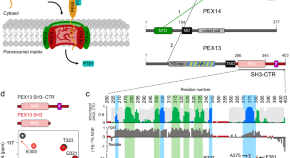
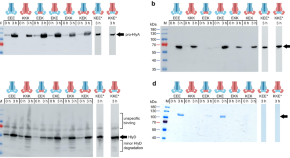
Modulation of peroxisomal import by the PEX13 SH3 domain and a proximal FxxxF binding motif
Import of proteins into peroxisomes depends on PEX5, PEX13 and PEX14. Here the authors obtain crystal structures and NMR data to show the recognition of diaromatic peptide motifs on a noncanonical surface of the PEX13 SH3 domain, revealing a dynamic network which modulates peroxisomal matrix import.
- Stefan Gaussmann
- Rebecca Peschel
- Michael Sattler
Cross-link assisted spatial proteomics to map sub-organelle proteomes and membrane protein topologies
The spatial mapping of proteins can give important functional insights. Here, Zhu et al. develop a cross-linking mass spectrometry-based spatial proteomics method that does not require protein engineering, affords sub-organelle resolution, and elucidates both protein locations and membrane topologies.
- Kerem Can Akkaya
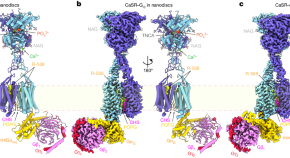
Promiscuous G-protein activation by the calcium-sensing receptor
Structures of the human calcium-sensing receptor can be bound into complex with G proteins from three different Gα subtypes while maintaining G-protein-binding specificity.
- Jinseo Park
- Qing R. Fan
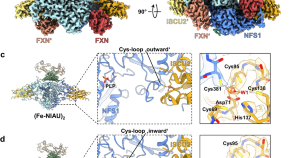
Mechanism and structural dynamics of sulfur transfer during de novo [2Fe-2S] cluster assembly on ISCU2
The biogenesis of iron-sulfur proteins in eukaryotes is initiated by the mitochondrial core ISC complex. Here, the authors provide structural, biochemical and spectroscopic data to characterize sulfur transfer intermediates in the core ISC complex.
- Vinzent Schulz
- Ralf Steinhilper
- Roland Lill
News and Comment
Shifting our perspective on orphan g protein-coupled receptors.
G protein-coupled receptors (GPCRs) with no known endogenous ligand are termed orphans. Deorphanization of a GPCR involves identifying the ligand, which can be a painstaking exercise. In this Comment, we discuss the challenges in the process, its role in drug discovery and alternative approaches to characterizing orphan GPCRs.
- Nicola J. Smith
- Fiona Murray
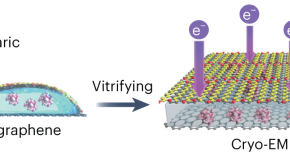
Graphene sandwich for cryo-EM
- Arunima Singh

The regulatory landscape of chromatin accessibility
A study in Nature Genetics identifies many regulators of genome-wide chromatin accessibility and then reports the mechanistic underpinnings for one of the identified transcription factors.
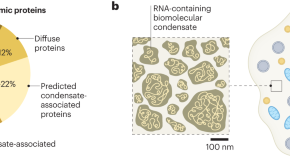
Mesoscale condensates organize the cytoplasm
Biomolecular condensates are recognized for their ability to compartmentalize the cytoplasm without bounding membranes, but the degree to which they organize the cytoplasm has not been clear. A new study reveals that condensates at a scale of 100 nm are responsible for the organization of at least 18% of the cytoplasmic proteome.
- Leshani Ahangama Liyanage
- Jonathon A. Ditlev
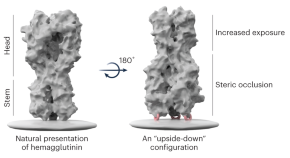
Turning vaccine design on its head
- Katarzyna Ciazynska

Micropolarized to the core
Macromolecules can undergo liquid–liquid phase separation to form condensates that have critical roles in biological functions and dysfunctions. A new study demonstrates that differences in micropolarity between components is a prime determinant of the multiphasic architecture of biomolecular condensates.
- My Diem Quan
- Josephine C. Ferreon
- Allan Chris M. Ferreon
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies

Earth Day 2024 is a mix of woke and political correctness, so the Earth gets a raw deal
"Planet vs Plastics" is the theme of Earth Day this year. Why? Because, according to the event’s organizers, "plastics are a danger to humanity and all living creatures, disrupting the balance of life on Earth." Therefore, "EARTHDAY.ORG is unwavering in our commitment to end plastics for the sake of human and planetary health, demanding a 60% reduction in the production of ALL plastics by 2040."
That sort of hyperbolic condemnation is typical of many recent Earth Days. The reality is that Earthlings and plastics must coexist. In many ways, plastics improve our quality of life. They have revolutionized medicine with life-saving devices, made space travel possible, lightened cars and planes (conserving fuel and lowering air pollution), and made all manner of consumer goods less expensive.
It is true that their convenience and low cost have spawned a throw-away culture that has a dark side: Single-use plastics currently account for about 40% of the plastic produced every year. That percentage could and should be reduced, but the absolutism of the Earth Day organizers illustrates what is wrong with today’s environmental movement.
HALF OF US STATES REVOLT AGAINST EPA CRACKDOWN ON GAS CARS WITH MAJOR LEGAL CHALLENGE
The first Earth Day celebration, a nationwide environmental teach-in held in 1970, was the brainchild of Democratic Senator Gaylord Nelson of Wisconsin, who was interested in environmental issues.
He recruited Pete McCloskey, a conservation-minded liberal Republican California congressman, to serve as his co-chair. In the spirit of the time, it was a touchy-feely, consciousness-raising New Age experience, and most activities were organized at the grassroots level.
READ ON THE FOX NEWS APP
Sadly, today’s Earth Day shares much with the current zeitgeist: It reeks of wokeness, political correctness and virtue signaling. It has devolved into an occasion for environmental activists to prophesy apocalypse, dish anti-technology dirt and allow passion and zeal to trump reality.
A generation ago, the mantra was simple and broadly accepted: "reduce, re-use and recycle." However, we hasten to note, as John Tierney highlighted in his classic New York Times Magazine piece, "Recycling is Garbage," recycling isn’t always beneficial.
Instead of setting up a false dichotomy of "Planet vs. Plastics," environmental activists would do well to redirect their zeal and predictions of catastrophe to imagining what we can do to protect the planet. A little innovation can go a long way toward protecting our environment.
Yet, unfortunately, many of those stumping for Earth Day on April 22 this year will oppose environment-friendly advances in science and technology. Consider these several examples:
We are now scratching the surface for ingenious ways to remediate contaminated soil by using plants with the ability to remove toxins such as heavy metals from soil. A comprehensive review of the progress and promise of this technique, known as phytoremediation, states that, "Plant abilities to uptake, translocate, and transform heavy metals, as well as to limit their toxicity, may be significantly enhanced via genetic engineering."
However, most prominent environmental groups oppose genetic engineering and would rather soil a promising technology than abandon a deeply held ideological position.
Similarly, some green groups opposed one of the most innovative efforts to address allegations of climate change caused by animal agriculture – the Impossible Burger, an ingredient of which is a plant-sourced protein, leghemoglobin.
The protein contains heme, also present in animal meat, and is partly responsible for the taste, texture and appearance of bloody-good meat. As Business Insider noted, "a handful of environmental activists have also taken issue with the burger. But their issue with the burger isn't heme — it's the fact that the Impossible Burger is made using genetically engineered ingredients, or GMOs. Those concerns largely take the shape of the old and unsubstantiated claim that GMOs cause everything from autism to cancer, despite the scientific consensus that they are safe."
A pervasive Earth Day meta-message will be disdain for the capitalist system that drives the innovation needed for effective environmental protection and conservation. (It’s no coincidence that low-income countries tend to be the most polluted.)
"Education" features prominently in Earth Day activities, as in, "Fifty four years ago, the first Earth Day started an environmental revolution. Now, we are igniting an education revolution to save the planet. EARTHDAY.ORG passionately envisions a world where every student ... is embraced by a rich education weaving together climate and environmental understanding, coupled with a strong civic engagement component."
Well, education can take many forms. For a previous Earth Day, seventh graders at a tony private school near San Francisco were given an unusual Earth Day assignment: Make a list of environmental projects that could be accomplished with Bill Gates’s fortune.
This approach to environmental awareness fits in well with the progressive worldview that the right to private property is subsidiary to undertakings that enlightened thinkers deem worthwhile.
And how interesting that the resources made "available" for the students’ thought experiment were not, say, the aggregate net worth of the members of Congress but the wealth of one of the nation’s most successful and most innovative entrepreneurs.
CLICK HERE FOR MORE FOX NEWS OPINION
Another Earth Day assignment for those same students was to read Rachel Carson’s best-selling 1962 book, "Silent Spring," an emotionally charged but deeply flawed excoriation of the widespread spraying of chemical pesticides to control insects.
As described by Roger Meiners and Andy Morriss in their scholarly yet eminently readable 2012 analysis, "Silent Spring at 50: Reflections on an Environmental Classic," Carson exploited her reputation as a well-known nature writer to advocate and legitimatize "positions linked to a darker tradition in American environmental thinking: neo-Malthusian population control and anti-technology efforts."
Carson’s proselytizing and advocacy led to the virtual banning of DDT and restrictions on other chemical pesticides even though "Silent Spring" was replete with gross misrepresentations and atrocious scholarship.
Her observations about DDT were meticulously rebutted point by point by J. Gordon Edwards, a professor of entomology at San Jose State University, a longtime member of the Sierra Club and the Audubon Society, and a fellow of the California Academy of Sciences.
In his stunning 1992 essay, "The Lies of Rachel Carson," Edwards demolished her arguments and assertions and called attention to critical omissions, faulty assumptions, and outright fabrications.
Meiners and Morriss concluded correctly that the influence of "Silent Spring" "encourages some of the most destructive strains within environmentalism: alarmism, technophobia, failure to consider the costs and benefits of alternatives, and the discounting of human well-being around the world." Sounds a lot like the agenda of Earth Day 2024.
Henry Miller, a physician and molecular biologist, is the Glenn Swogger Distinguished Fellow at the American Council on Science and Health. He was a research associate at the National Institutes for Health and the founding director of the FDA’s Office of Biotechnology. Follow them on Twitter at @henryimiller.
Original article source: Earth Day 2024 is a mix of woke and political correctness, so the Earth gets a raw deal


IMAGES
VIDEO
COMMENTS
protein, highly complex substance that is present in all living organisms.Proteins are of great nutritional value and are directly involved in the chemical processes essential for life.The importance of proteins was recognized by chemists in the early 19th century, including Swedish chemist Jöns Jacob Berzelius, who in 1838 coined the term protein, a word derived from the Greek prōteios ...
a protein's shape determines its function. Conversely, DNA has just one function—genetic information storage—for which it assumes a single shape. The sequence of amino acids in a protein determines its folded structure The specific order of amino acids in a protein is known as its primary structure. It is this sequence that determines the ...
Proteins and Polypeptides. Proteins are organic compounds that contain four elements: nitrogen, carbon, hydrogen, and oxygen. To comprehend the full scope of proteins, it is crucial to understand various properties, including the basic biological molecule, peptides, polypeptide chains, amino acids, protein structures, and the processes of protein denaturation.
For instance, the blood protein hemogobin is made up of four polypeptide chains, each of which also contains a heme molecule, which is ring structure with an iron atom in its center. Proteins have different shapes and molecular weights, depending on the amino acid sequence. For example, hemoglobin is a globular protein, which means it folds ...
Hormones are chemical-signaling molecules, usually small proteins or steroids, secreted by endocrine cells that act to control or regulate specific physiological processes, including growth, development, metabolism, and reproduction. For example, insulin is a protein hormone that helps to regulate the blood glucose level.
Proteins are organic molecules that are present in living organisms. They serve a wide range of functions including organization, transportation, and defense. Proteins are composed of amino acid chains, and structure levels are up to four. Certain specific protein examples include collagen, insulin, and anticorps. Q4.
Amino acids. Amino acids are the monomers that make up proteins. Specifically, a protein is made up of one or more linear chains of amino acids, each of which is called a polypeptide. (We'll see where this name comes from a little further down the page.) There are 20 types of amino acids commonly found in proteins.
Amino Acid Structure. Amino acids are the monomers that make up proteins. Each amino acid has the same core structure, which consists of a central carbon atom, also known as the alpha (α) carbon, bonded to an amino group (NH2), a carboxyl group (COOH), and a hydrogen atom. Every amino acid also has another atom or group of atoms bonded to the ...
The first of these is the primary structure, which is the number and sequence of amino acids in a protein's polypeptide chain or chains, beginning with the free amino group and maintained by the peptide bonds connecting each amino acid to the next. The primary structure of insulin, composed of 51 amino acids, is shown in Figure 16.5.1 16.5. 1.
This page contains materials for the biochemistry class session on protein structure. It features a 1-hour lecture video, and also presents the prerequisites, learning objectives, reading assignment, lecture slides, homework with solutions, and resources for further study.
It is estimated that the human body contains well over a million different kinds of protein, and even a single-cell organism contains thousands. Each of these is a polymer of amino acids which has a highly specific composition, a unique molecular weight (usually in the range from 6000 to 1 000 000) and its own sequence of different amino acids ...
Protein Denaturation. Protein denaturation is the process of destruction of the quaternary, tertiary, and secondary structure of proteins by the application of external force or strong chemicals like acid and base. Denaturation of protein might destroy cell activity as most of the metabolic pathways require proteins in one form or the other.
9.5: Protein Structure Proteins may be fibrous, or globular. A protein may have up to four levels of structure. 9.6: Denaturation of Proteins A wide variety of reagents and conditions can cause a protein to denture. Denaturation leads to loss of biological activity of the protein due to loss of three-dimensional structure. 9.7: Digestion of ...
These are of following types:-. Nucleoproteins: Combination of protein and nucleic acid. Mucoproteins: Combination of proteins and carbohydrates (>4%) Glycoproteins: Combination of proteins and carbohydrates (<4%) Chromoproteins: Combination of proteins and coloured pigments. Lipoproteins: Combination of proteins and lipids.
Assignments for core residues, which are important for the protein structure, are generally more accurate than for the entire protein, in particular for core Ala, Cys, and Asp residues, which show ...
Proteins questions. A new drug is developed which selectively cleaves covalent bonds between two sulfur atoms of non-adjacent amino acids in a polypeptide chain. Which level of protein structure in affected molecules would be most directly affected by the drug?
A protein's structure is primarily made up of long chains of amino acids. The arrangement and placement of amino acids give proteins certain characteristics. All amino acid molecules contain an amino (-NH2) and a carboxyl (-COOH) functional group. Hence, the name "Amino-Acid". Polypeptide chains are synthesized by linking together amino ...
The translation is the second part of the central dogma of molecular biology: RNA --> Protein. It is the process in which the genetic code in mRNA is read to make a protein. The translation is illustrated in Figure 6.4.6. After mRNA leaves the nucleus, it moves to a ribosome, which consists of rRNA and proteins.
Definition. Protein synthesis is process in which polypeptide chains are formed from coded combinations of single amino acids inside the cell. The synthesis of new polypeptides requires a coded sequence, enzymes, and messenger, ribosomal, and transfer ribonucleic acids (RNAs). Protein synthesis takes place within the nucleus and ribosomes of a ...
Knowledge about protein structure assignment enriches the structural and functional understanding of proteins. Accurate and reliable structure assignment data is crucial for secondary structure prediction systems. Since the 1980s, various methods based on hydrogen bond analysis and atomic coordinate geometry, followed by machine learning, have been employed in protein structure assignment ...
Four levels of structure of proteins. The principal, secondary, tertiary and quaternary levels of protein structure are the four stages. To fully understand how a protein functions, it is helpful to understand the purpose and role of each level of protein structure. Q3.
Description: This lecture on predicting protein structure covers refining a partially correct structure. Methods include energy minimization, molecular dynamics, and simulated annealing. ... assignment_turned_in Programming Assignments with Examples. assignment Presentation Assignments. assignment Written Assignments. co_present Instructor ...
Proteins are biopolymers of amino acids (polypeptides), joined by peptide bonds, that are generated by ribosomes. The amino acid sequence, encoded by its gene, determines a protein's structure ...
The protein contains heme, also present in animal meat, and is partly responsible for the taste, texture and appearance of bloody-good meat. ... Another Earth Day assignment for those same ...